Understanding Ductal Carcinoma In Situ
 |
|
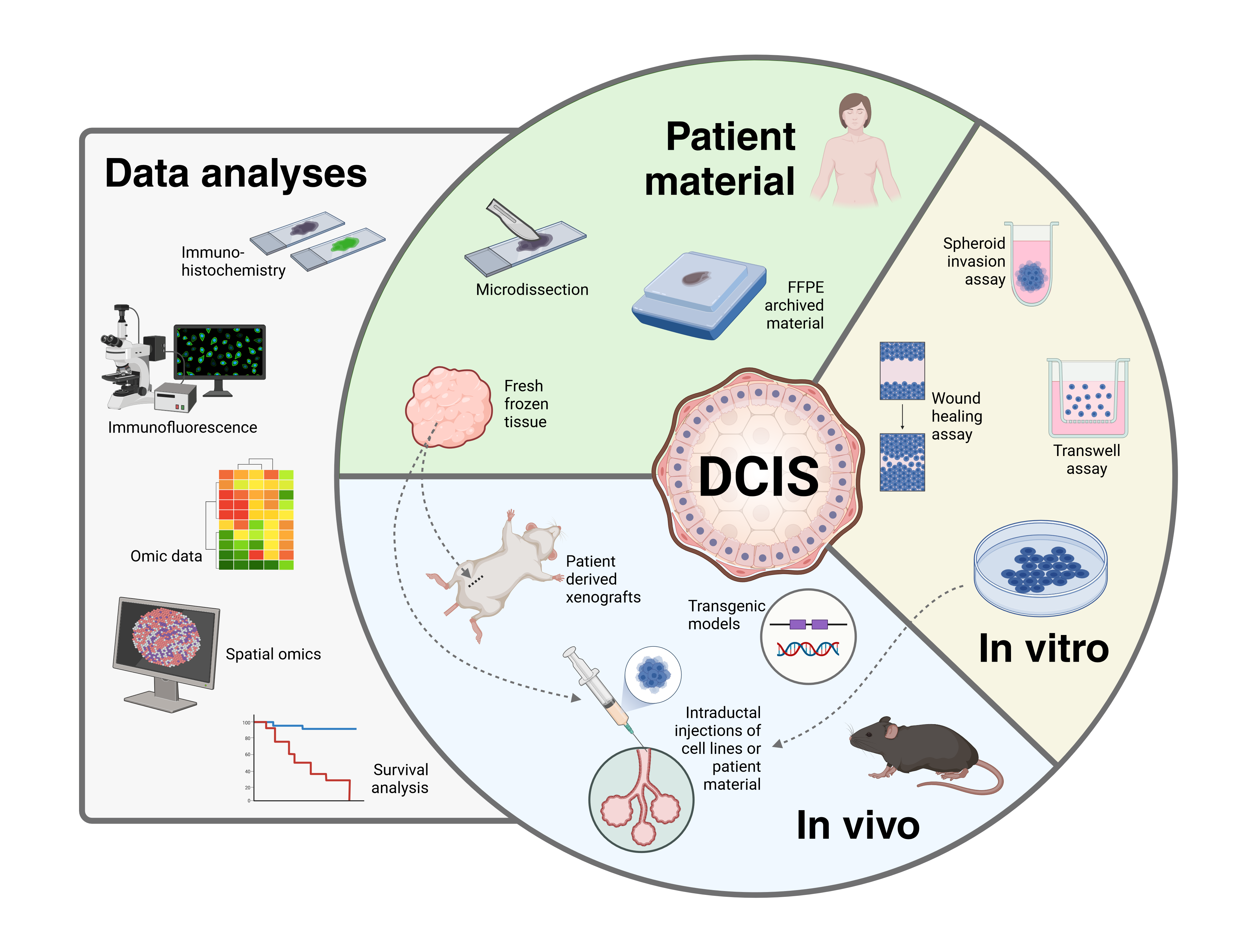
Our group has extensive experience working with DCIS and currently we have three DCIS-related projects.
Participants: Helga Bergholtz, Jens Henrik Norum, Phuong Vu, Helene Midtun Flatekvål, Karin Teien Lande
Multi-level Characterization of Ductal Carcinoma In Situ (DCIS) to improve risk stratification and treatment options
Our group is part of the OUH - Centre for breast cancer. We are prospectively collecting tissue and blood samples from patients undergoing surgery for DCIS at Oslo University Hospital. The signed consent forms are digitalized and stored in MedInsight. For more information contact the project leader Therese Sørlie (tsorlie@ous-hf.no).
Unraveling the ambiguous role of HER2 in DCIS and breast cancer progression
Funded by Pink Ribbon, Norwegian Cancer Society
The prevalence of HER2-positive lesions is higher in DCIS than in IDC. This apparent predilection towards non-invasive growth contrasts the aggressive phenotype of HER2-positive invasive breast tumors. In this project we aim to elucidate the role of HER2 in breast tumor invasion. We use genomic data and tissue samples from patients with DCIS or invasive breast cancer and breast cancer xenografts. The project is divided into two work packages:
In vitro and in vivo modeling of HER2-positive breast cancer progression and invasion. Here we use sophisticated intraductal xenografts and in vitro invasion assays with human and murine cell lines to model intraductal tumor growth and invasion.
Mapping the molecular landscape of HER2-positive human breast cancer progression. We have access to multiomic datasets including data from DCIS and invasive tumors from patients that enables us to explore molecular characterization. In addition, we perform Spatial transcriptomic analyses on selected samples to explore the interaction between tumor cells and stroma in HER2-positive tumors.
Spatial heterogeneity and breast cancer lineages in Ductal Carcinoma In Situ
Funded by the Norwegian Cancer Society
In this project we aim to clarify the biological importance of the two main subtypes of breast cancer, luminal A and basal-like, in DCIS and investigate the impact of the tumor microenvironment on the subtypes and on tumor progression. We seek insights into the spatial molecular heterogeneity within tumor cells and within the microenvironment surrounding the ducts and how this may impact tumor progression.
Spatial transcriptomics of DCIS tumor and microenvironmental cell populations. We use digital spatial profiling (DSP) for high-plexed molecular profiling of tissues and subsequent digital quantitation of target analytes. Our cohort includes >470 DCIS cases operated during 1996-2018.
Rate of tumor progression in luminal vs. basal-like mouse models of breast cancer. We use the Mouse mammary IntraDuctal (MIND) method to address the properties of cell growing intraductally and their potential to invade the surrounding adipose tissue. These studies include immune defect, immune competent and genetically modified models.
Busy Bs - The role of B cells in breast tumor invasion
Funded by South-Eastern Norway Regional Health Authority
Like bees are attracted to a pot of honey, lymphocytes are attracted to tumor cells. In breast ductal carcinoma in situ (DCIS), tumor cells are confined within the milk ducts without invading the surrounding tissue. Still, immune cells are attracted to the tumor cell-filled ducts and accumulation of lymphocytes around DCIS lesions is common.
The immune cells that surround DCIS might hold the key to understanding what drives invasion in breast cancer and thus enable discovery of new biomarkers for progression. In this project we will investigate the immune microenvironment in DCIS and its role in invasion with special emphasis on B cells and explore what attracts B cells and other immune cells to tumor-filled ducts; the “honey” of DCIS.
We will use a sophisticated in vivo method: Mouse INtraDuctal injections (MIND), to model intraductal growth and invasion in immunocompetent mice. Using murine cell lines of different molecular subtypes, we aim to disentangle the differences in immune response between the subtypes. To explore specifically the role of B cells in the invasion process, we will perform B cell depletion using CD20 monoclonal antibodies. In addition, we will study the spatial context of B cell infiltration in the invasion process using state-of-the-art spatial proteomic/transcriptomic platforms and in silico cell deconvolution methods on transcriptomic data from bulk tumor tissue samples.
Relevant publications:
- Bergholtz, H. et al. B cells and energy metabolism in HER2-positive DCIS: insights into breast cancer progression from spatial-omics analyses. Breast Cancer Research 27, 44 (2025).
- Moragas, N. et al The SEMA3F-NRP1/NRP2 axis is a key factor in the acquisition of invasive traits in in situ breast ductal carcinoma. Breast Cancer Research 26, 122 (2024)
- Langille, E. et al. Loss of Epigenetic Regulation Disrupts Lineage Integrity, Induces Aberrant Alveogenesis, and Promotes Breast Cancer. Cancer Discovery 12, 2930–2953 (2022).
- Xu, H. et al. Multi-Omics Marker Analysis Enables Early Prediction of Breast Tumor Progression. Frontiers in Genetics 12, (2021).
- Bergholtz, H. et al. Comparable cancer-relevant mutation profiles in synchronous ductal carcinoma in situ and invasive breast cancer. Cancer Reports 3, (2020).
- Bergholtz, H. Deciphering molecular heterogeneity and relevance of subtypes in breast cancer progression. (2020). Doctoral thesis.
- Bergholtz, H. et al. Contrasting DCIS and invasive breast cancer by subtype suggests basal-like DCIS as distinct lesions. npj Breast Cancer 6, 1–9 (2020).
- Lesurf, R. et al. Molecular Features of Subtype-Specific Progression from Ductal Carcinoma In Situ to Invasive Breast Cancer. Cell Reports 16, 1166–1179 (2016).
- Borgquist, S. et al. The prognostic role of HER2 expression in ductal breast carcinoma in situ (DCIS); a population-based cohort study. BMC cancer 15, 468–468 (2015).
- Zhou, W. et al. Molecular subtypes in ductal carcinoma in situ of the breast and their relation to prognosis: a population-based cohort study. BMC cancer 13, 512–512 (2013).
- Kristensen, V. N. et al. Integrated molecular profiles of invasive breast tumors and ductal carcinoma in situ (DCIS) reveal differential vascular and interleukin signaling. Proc Natl Acad Sci U S A 109, 2802–2807 (2012).
- Muggerud, A. A. et al. Molecular diversity in ductal carcinoma in situ (DCIS) and early invasive breast cancer. Molecular oncology 4, 357–368 (2010).
- Muggerud A. A. et al. Frequent aberrant DNA methylation of ABCB1, FOXC1, PPP2R2B and PTEN in ductal carcinoma in situ and early invasive breast cancer. Breast Cancer Res 12, R3 (2010).
- Zhou, W. et al. Full sequencing of TP53 identifies identical mutations within in situ and invasive components in breast cancer suggesting clonal evolution. Molecular oncology 3, 214–219 (2009).
