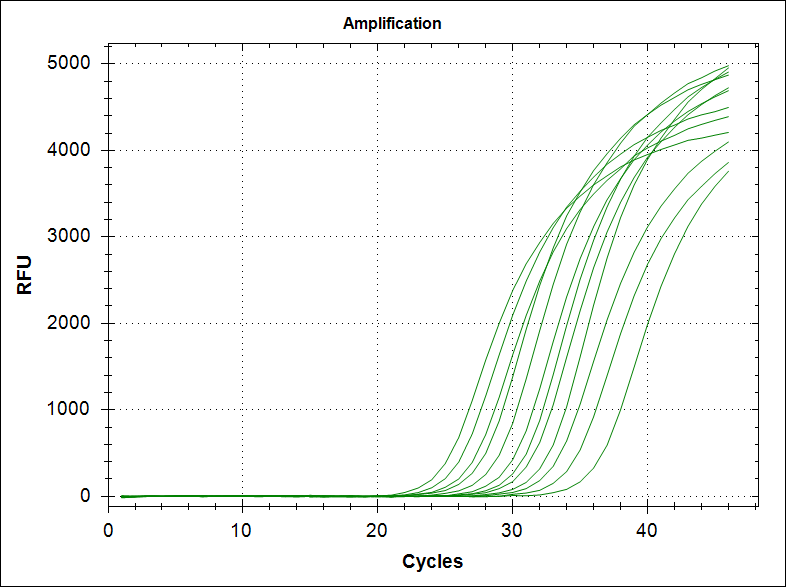
Material and recipes
The Sars-Cov-2 virus genome is a 29903 base pair linear sequence of RNA. Thus, to amplify the fragments by real-time PCR two enzymes is needed. One enzyme (reverse transcriptase) to create a template of cDNA, and the second enzyme (DNA polymerase) for the PCR amplification. If TaqMAN probes are used in the real-time PCR, the enzyme needs to have a 5’ to3’ prime exonuclease activity. The enzymes used was cloned, expressed (in bacteria) and purified by David J. Warren at the central laboratory. None of this work would have been possible without his expertise. From the first test production of enzymes we obtain some 2x106 units of Taq and 4x105 units of reverse transcriptase MasnUp. The plasmids used to produce the two different enzymes were kindly donated to us by Pipette Jockey. The weekly testing of chemistry was performed by Julia Elisabeth Simensen.
Chemistry
Initially many different buffers were tested based on the literature. However, the buffer that worked best in our lab was a homemade regular Thermopol buffer (with or without MgSO4 in the 10X). The component of a 10X Thermopol buffer (without MgSO4) is the following; 200mM Tris-HCl, 100mM (NH4)2SO4, 100mM KCl, 1% Triton® X-100 with pH 8.8 @ 25°C.
Stock solution of MgSO4 and BSA was made to 200mM and 10mg/ml, respectively.
Primers from the table were order from Integrated DNA Technologies IDT.
For real-time PCR, without TaqMAN probes, we used Eva Green as a reporter dye. This dye is superior to Syber Green in our mixes.
dNTPs were mixed (final concentration of 25mM of each dNTP) in stock solutions of 100mM and kept frozen. Re freezing and thawing will degrade the dNTP with the result of poor PCR.
Sterile water of i.v. injection quality was used in the PCRs.
Control virus RNA was obtained from three different sources, IDTdna, 2019-nCoV_N Positive Control, SARS-CoV Control and MERS-CoV Control, Twist Synthetic, SARS-CoV-2 RNA control 1, SARS-CoV-2 RNA control 1 and from Magnar Bjørås. Additionally, we were also able to get some “real” RNA sample from a Covid-19 testing lab at OUS.
Commercial qScript cDNA synthesis kit and perfecta qPCR super mix (Quanta Biosciences) was used as comparison to in-house chemistry.
Taq's optimum temperature for activity is 75–80 °C, but has activity at lower temperatures. This could potentially affect a one tube chemistry of reverse transcription and real-time PCR. Thus, hybridoma the produce anti bodies against Taq was acquired from ATCC. We found that the ratio 1µg antibodies/5 unit of Taq gave sufficient inhibition of Taq at low temperatures. And that the effect of the antibodies was removed by an initial denaturation step.
Example of real-time PCR recipe with homemade chemistry. Starting material, 1µl cDNA from a separate reaction, diluted in a 2-fold series. Cycling condition, initial denaturation 94°C 1 minute, then 45 cycles with 94°C 20 sec, 56°C 20sec and 72°C 40 sec.
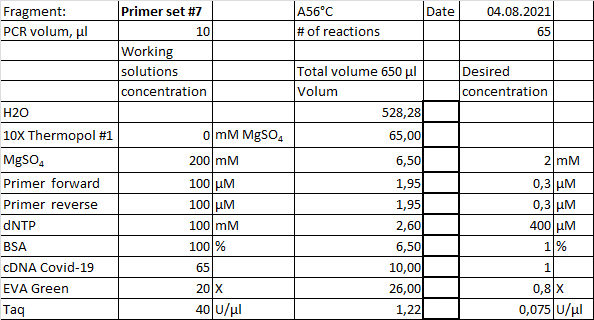
Amplification curves