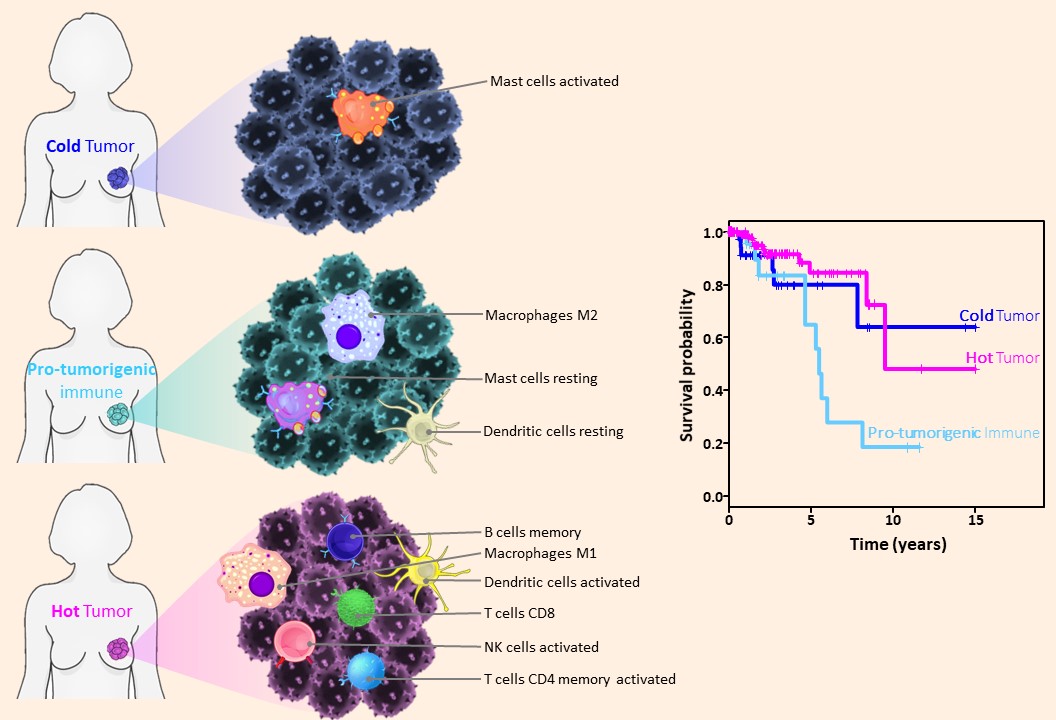
Breast cancer immune clusters.
In 2019, we discovered a new breast cancer subtype: immune clusters, (Tekpli et al., Nature Communications, 2019), we clearly show how the information on immune infiltration in breast cancer is clinically applicable and relevant for treatment decision.
We invariably identified a group of patients with a poor prognosis (see figure below) and poor response to treatment: this group of breast cancers needs new personalized therapies.
We therefore researching on
- developing an assay to efficiently subtype breast cancer patients for immune clusters.
- identify test and validate new treatment strategies in pre-clinical setting.
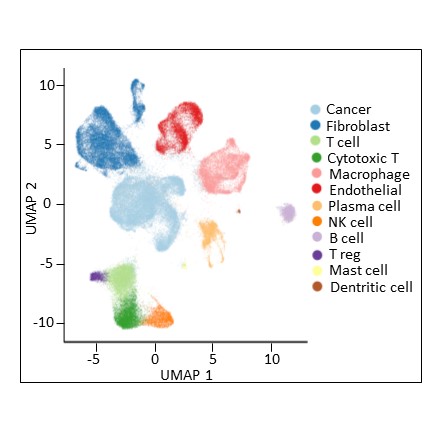
Single cell analysis of breast tumors at different time points of therapies
We are analyzing biopsies, before, between and at the end of treatment for 3 clinical trials:
NeoLetExe: A trial assessing the effect of aromatase inhibitors (letrozole and exemestane) in postmenopausal, locally advanced patients with ER+ breast cancer.
NeoLetRib: Patients with locally advanced, ER-positive cancer receive neoadjuvant therapy with letrozole (Femar) in combination with ribociclib (Kisquali) for 6 months.
ALICE: A randomized phase IIb study evaluating the effects of the combination of chemotherapy and immunotherapy in triple negative metastatic breast cancers.
The main method applied on these biopsies is single cell RNA sequencing on freshly dissociated cells. We also aim in the future at using the remaining cryopreserved material from these biopsies to apply, in collaboration, one or several of the following: scATAC-seq, scDNA-seq, scChIP-seq.
Spatial mapping of tumor, immune and stromal cells: spatial transcriptomic
Different cell types in breast tumors interact and 'communicate' to build local ecosystem in which cancer cells grow. We need to determine how the different cell types organize and interact spatially. We use different strategies from multiplexed immunohistochemistry to spatial transcriptomics methods.
- In collaboration with Dr. Alexandre Corthay’s lab, we use multiplexed immunostaining techniques to study in normal breast / tumor pairs, the location and infiltration of different immune cell types (see below an example of such immunostaining).
- To uncover the cellular heterogeneity in histological sections, we localize mRNA molecules directly on a tissue slide which allows us to understand tissue heterotypic interactions and tissue cellular ecosystems, for this, we use spatial transcriptomics (Visium method from 10X genomics).
- We also plan to use the Nanostring GeoMx method which will be available at Oslo University Hospital in fall 2021 to study in detail the immune and stromal tumor microenvironment of different breast cancer subtypes.
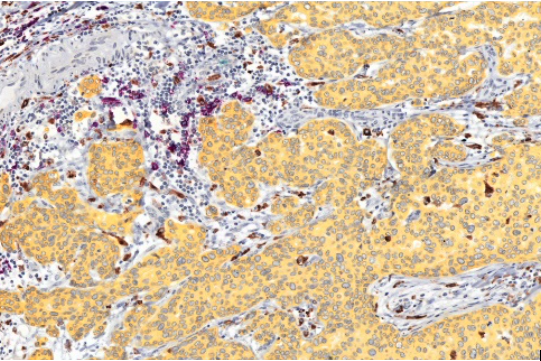
Purple: B cells Teal: Granulocytes Brown: macrophages |
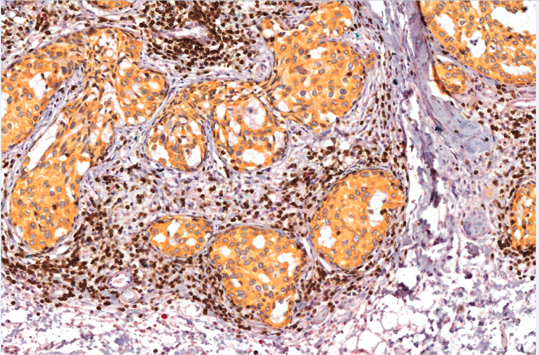
Red: NK cells Teal: Mast cells Brown: T cells (CD3) |
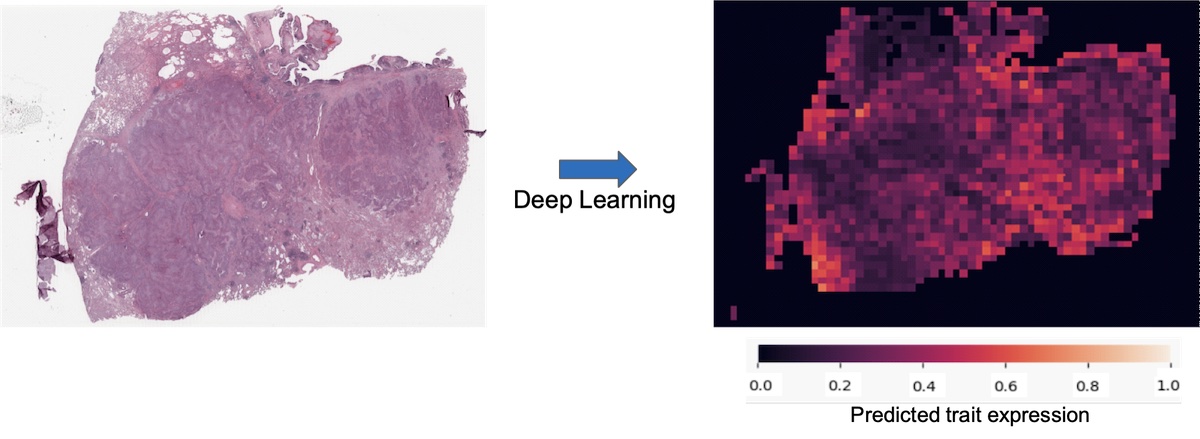
Digital pathology through machine learning analysis of breast cancer diagnostic slides.
Histopathological images are routinely produced for diagnosis and stratification of cancer. Image-based exercises are ideally suited for deep learning and are anticipated to provide significant time and cost-saving measures in precision oncology.
We hypothesize that machine learning algorithm can recognize important traits which could be helpful to guide diagnosis, treatment, or make a prognosis directly on the routinely performed H&E stain of breast tumors. In collaboration with Prof. Zohar Yakhini and Dr Øystein Garred, we have set up artificial neural network (a class of powerful machine learning methods), we train such machine learning algorithms to implement the field of digital pathology in breast cancer.
Patient-derived organoids at different time points of therapy
We derive organoids from the primary dissociated cells obtain from freshly resected breast tumors.
Patient-derived organoids are three-dimensional in vitro models, initially grown from a patient cancer cells. They mimic the phenotype of the primary tumor and several studies demonstrated that they recapitulate features of primary tumors, including histological complexity and genetic heterogeneity
We obtain these ex-vivo patient cultures from two clinical trials, the NeoLetExe and NeoLetRib, in which locally advanced ER+ breast cancer patients are treated in the neoadjuvant setting.
In our lab, when primary breast tumors biopsies are withdrawn and enzymatically digested, up to 25.000 cells are used for single cell RNA-seq. On the same day and with the same pool of dissociated cells, 50.000 – 150.000 cells are used to generate organoids.
We first aim at carefully biobanking such patient-derived organoids at different time point of therapy. We will also have the opportunity to test ex-vivo treatment strategies which we predict to be more efficient at the individual patient level based on the information obtained from single cell RNA sequencing.
