Parkinson genomics
Genetic research has a major role to play in the effort to elucidate the pathogenesis of PD on a molecular level. With respect to the sporadic, complex form of the disease, a major advance in recent years has been the identification of susceptibility loci through large-scale genome wide association studies (GWAS). A total of 26 loci have been associated with disease risk at genome-wide significance levels (Nalls, Nat Genet 2014). In addition, there is clear evidence that within several of these regions multiple genetic variants are independently increasing disease risk (unpublished data). However, considerable scientific efforts are necessary to refine and expand the understanding of GWAS association signals before the full potential value of these findings can be harvested for further research.
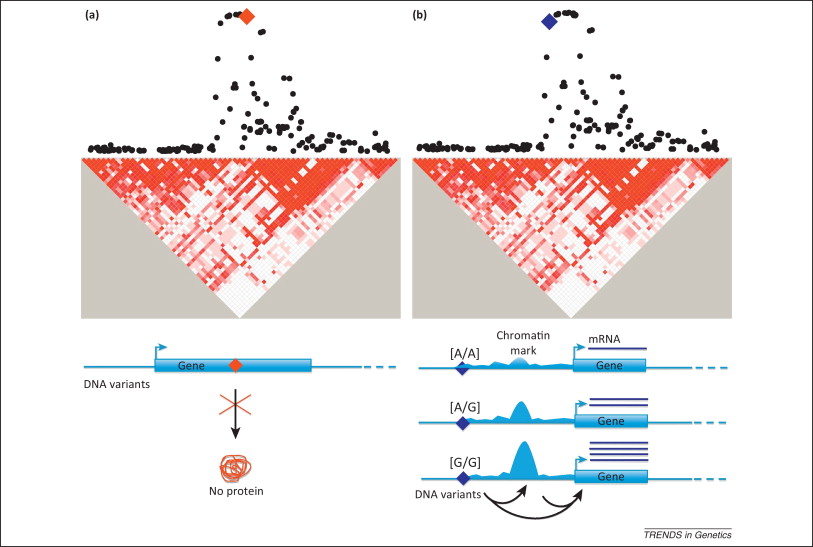
Genomic studies can identify genetic variants influencing disease risk using a combination of research tools and methods
Association signals emerging from GWAS typically involve dozens of gene variants in linkage disequilibrium (LD), meaning that they are inherited together more often than would be expected by chance. This phenomenon complicates the identification of the functionally relevant variants within risk loci. Furthermore, most genetic risk variants fall outside coding regions and do not alter the amino acid sequence of proteins (Abecasis, Nature 2012). Until recently, the functional characterization of risk-associated loci was hindered by the limited knowledge of the human genome outside coding sequences. However, approaches to successfully characterize the functional nature of these loci are emerging. A series of large-scale genomics projects, including the Encyclopedia of DNA Elements (ENCODE) and the International Human Epigenome Consortium (IHEC), are leading an effort to annotate the non-coding regions of the human genome. Together, these layers of biological information serve as the foundation for post-GWAS functional studies. Recent studies have indicated that the majority of genetic risk variants regulate gene expression through mechanisms involving transcription factor (TF) binding, chromatin looping, or epigenetic mechanisms as DNA methylation and miRNA recruitment.
Previous studies providing functional explanations of GWAS signals are generally scarce, yet a set of successfull early publications demonstrate that the task is feasible using a combination of targeted data mining and experimental follow-up (Edwards, Am J Hum Genet 2013). No study to date has systematically assessed GWAS signals in PD to nominate causal variants and follow-up functional hypothesis. Several more or less standardized methodological workflows for the functional exploration of GWAS loci have been published (Cooper, Nat Rev Genet 2011; Ward, Nat Biotechnol 2012; Edwards, Am J Hum Genet 2013; Ritchie, Genome Med 2014), yet available public data resources and computational tools are constantly evolving. As an example, pilot data from the Genotype-Tissue Expression (GTEx) Consortium were published May 2015, including eQTL data from substantia nigra and other specific brain regions highly relevant to PD (The GTEx Consortium, Science 2015). We will perform an up-to-date, comprehensive approach systematically exploiting existing public data to nominate causal variants within PD GWAS loci, followed by functional experiments to investigate mechanistic hypotheses.
Open positions: Scientia Fellowship (2-year postdoc), PhD position (3-years)
Funding: Research Council of Norway
