Standardization of methodology leads to innovation

The Epigenetics Group at the Department of Molecular Oncology, headed by Guro E. Lind, has developed a robust internal control for DNA methylation analyses by droplet digital PCR (ddPCR). The findings have recently been published in the journal Clinical Epigenetics. First author is Heidi Dietrichson Pharo.
The ddPCR technology (see Fig. 1) allows absolute quantification of nucleic acids, and has great potential for DNA methylation analyses of liquid biopsies. However, the lack of consensus regarding how to perform standardized methylation-specific ddPCR experiments has been challenging. The 4Plex internal control developed by Pharo et al. consists of four individual pericentromeric assays, and is analyzed in the same reaction as the target of interest. The 4Plex increases the precision of the analyses by a) reducing the variability in methylation concentrations, b) correcting for variable input amount, and c) reducing the effect of chromosomal aberrations (collaboration with the Lothe lab). PoDCall, an in-house developed algorithm for automated dichotomization of droplets, further contributes to consistency of the ddPCR results.
The work is an important contribution to the standardization of ddPCR-based methylation analyses, and is especially relevant for the rapidly evolving field of liquid biopsies, which holds great promise for precision medicine. It has resulted in two recently submitted patent applications, one regarding the 4Plex control, and the other regarding the PoDCall algorithm.
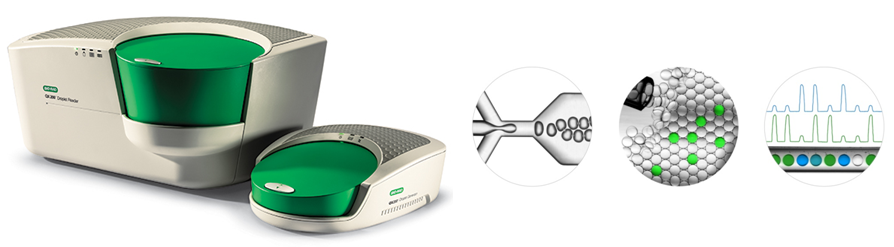
Fig. 1: The droplet digital PCR (ddPCR) platform from BioRad. With the ddPCR platform from BioRad, a PCR mixture is randomly divided into ~20,000 nanoliter-scale droplets formed by water-in-oil emulsion. Individual PCRs are performed inside each droplet, and based on the fraction of fluorescence-positive droplets, i.e. containing the target, absolute target quantification can be performed.
Links:
The article in Clinical Epigenetics:
A robust internal control for high-precision DNA methylation analyses by droplet digital PCR.
Pharo HD, Andresen K, Berg KCG, Lothe RA, Jeanmougin M, Lind GE.
Clin Epigenetics. 2018 Feb 21;10:24. doi: 10.1186/s13148-018-0456-5. eCollection 2018.
PMID: 29484034
