Combined genomics and transcriptomics in rare neurometabolic disease research
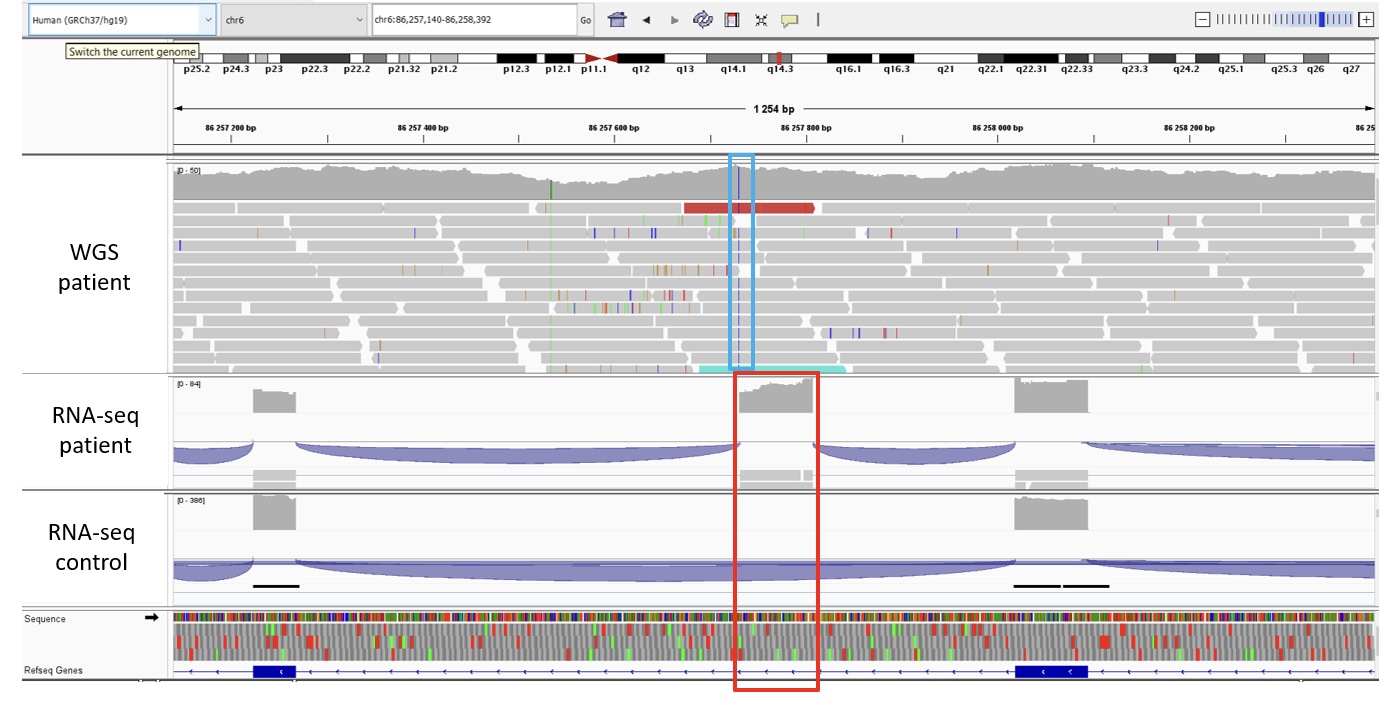
In a recently initiated project, we use RNA sequencing (RNA-seq) as a tool complementary to WGS-data analysis to identify disease causing variants whose clinical significance could be missed in standard WGS-data analysis, for example synonymous variants, intronic variants, intragenic deletions/duplications. Transcriptome analysis provides a shortcut to directly reveal consequences of genetic variants that result in aberrant gene expression levels or aberrant splicing or monoallelic expression. The bioinformatic analysis of RNA-seq data are performed using DROP and FRASER. One example that illustrates the approach is the identification of aberrant splicing in a patient that was diagnosis-negative after WGS-based analyses. RNA-seq revealed a pseudo-exon inclusion in SNX14 caused by an intronic variant identified by retrospective examination of this gene in the WGS data (see Figure).
These bioinformatics tools are now used to identify and characterize the disease etiology in patients with clinical symptoms of neurotransmitter disease. The children recruited to the project manifest with movement disorders and were without a genetic diagnosis after diagnostic analysis of WGS-data from parent-patient trios. The disorders studied in this project present predominantly with neurological features, including dystonic or dyskinetic cerebral palsy and movement disorders in general. Genetic conditions that lead to secondary defects in the synthesis, catabolism, transport and metabolism can lead to neurotransmitter abnormalities, which can present with similar features. These secondary defects are not well studied and finding the cause of the secondary neurotransmitter defect in more patients will give new insight into cell biology and the pathophysiology involved in severe neurometabolic diseases.
The project receives financial support by the Nasjonal kompetansetjeneste for sjeldne diagnoser (Norwegian National Advisory Unit on Rare Disorders, NKSD), Norway. 
Collaborators
Dr. L. Mathisen (AMG), Dr. C.L. Salvador (Dept. of Medical Biochemistry, OUS), and Prof. P. Aden, Dr. M. Oppebøen and Prof. P. Strømme (Division of Pediatric and Adolescent Medicine, OUS): patient recruitment, counseling and clinical investigations.
Dr. A. Sundaram and Dr. P.M. Bjørnstad (AMG): bioinformatics analyses. RNA-seq and WGS bioinformatics pipelines.
