Tumor heterogeneity of colorectal cancer
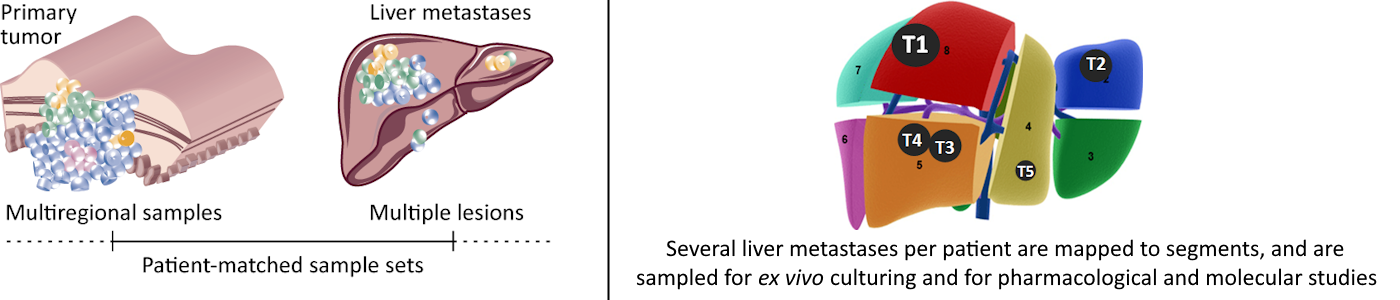
We aim to uncover the clinical consequences of spatio-temporal tumor heterogeneity of primary and metastatic colorectal cancers. We use “multi-omics” technologies and data integration to map molecular tumor profiles in the context of tumor heterogeneity. Spatial heterogeneity is analyzed among multiregional samples of each tumor and/or multiple metastatic lesions from each patient. Metastatic evolution is analyzed by comparisons of patient-matched primary tumors and metastases.
Computational modeling is used to reconstruct tumor architectures and evolution on a subclonal level and based on mutational patterns. Phenotypic plasticity is modeled based on gene expression programs and classifications. Tumor organoids cultured from the corresponding samples are used to model heterogeneity on the functional level and of drug sensitivities.
Selected publications:
Langerud J, Eilertsen IA, Moosavi SH, Klokkerud SM, Backe I, Hektoen M, Sjo O, Jeanmougin M, Tejpar S, Nesbakken A, Lothe RA, Sveen A. Multiregional transcriptomics identifies congruent consensus molecular subtypes with prognostic value beyond tumor heterogeneity in colorectal cancer. Nat Commun. 2024; 15(1):4342.
Sveen A, Johannessen B, Klokkerud SM, Kraggerud SM, Meza-Zepeda LA, Bjørnslett M, Bischof K, Myklebost O, Taskén K, Skotheim RI, Dørum A, Davidson B, Lothe RA. Evolutionary mode and timing of dissemination of high-grade serous carcinomas. JCI Insight 2024;9(3):e170423..
Bergsland CH, Jeanmougin M, Moosavi SH, Svindland A, Bruun J, Nesbakken A, Sveen A, Lothe RA. Spatial analysis and CD25-expression identify regulatory T cells as predictors of a poor prognosis in colorectal cancer. Mod Pathol. 35(9):1236-1246, 2022.
Moosavi SH, Eide PW, Eilertsen IA, Brunsell TH, Berg KCG, Røsok BI, Brudvik KW, Bjørnbeth BA, Guren MG, Nesbakken A, Lothe RA, Sveen A. De novo transcriptomic subtyping of colorectal cancer liver metastases in the context of tumor heterogeneity. Genome Med;13:143, 2021. DOI 10.1186/s13073-021-00956-1.
Berg KCG, Brunsell TH, Sveen A, Alagaratnam S, Bjørnslett M, Hektoen M, Brudvik KW, Røsok BI, Bjørnbeth BA, Nesbakken A, Lothe RA. Genomic and prognostic heterogeneity among RAS/BRAFV600E /TP53 co-mutated resectable colorectal liver metastases. Mol Oncol. 15(4):830-845, 2021.
Bruun J, Kryeziu K, Eide PW, Moosavi SH, Eilertsen IA, Langerud J, Røsok B, Totland MZ, Brunsell TH, Pellinen T, Saarela J, Bergsland CH, Palmer HG, Brudvik KW, Guren T, Dienstmann R, Guren MG, Nesbakken A, Bjørnbeth BA, Sveen A, Lothe RA. Patient-Derived Organoids from Multiple Colorectal Cancer Liver Metastases Reveal Moderate Intra-patient Pharmacotranscriptomic Heterogeneity. Clin Cancer Res. 26(15):4107-4119, 2020.
Sveen A, Løes IM, Alagaratnam S, Nilsen G, Høland M, Lingjærde OC, Sorbye H, Berg KC, Horn A, Angelsen JH, Knappskog S, Lønning PE, Lothe RA. Intra-patient inter-metastatic genetic heterogeneity in colorectal cancer as a key determinant of survival after curative liver resection. PLoS Genet 12:e1006225, 2016. DOI 10.1371/journal.pgen.1006225.
Giercksky HE, Thorstensen L, Qvist H, Nesland JM, Lothe RA. Comparison of genetic changes in frozen biopsies and microdissected archival material from the same colorectal liver metastases. Diagn Mol Pathol. 6(6):318-25, 1997.
