Structures of Endonuclease V in complex with DNA reveal molecular basis for repair of deaminated bases in DNA.

Bjørn Dalhus
In a recent paper by Dalhus et al., published in Nature Structural and Molecular Biology (impact factor 11.085), scientists at Institute for Medical Microbiology, Section for Molecular Biology and Institute for Clinical Biochemistry published structures of DNA repair enzyme Endonuclease V in complex with DNA. The work was carried out in the lab of Magnar Bjørås.
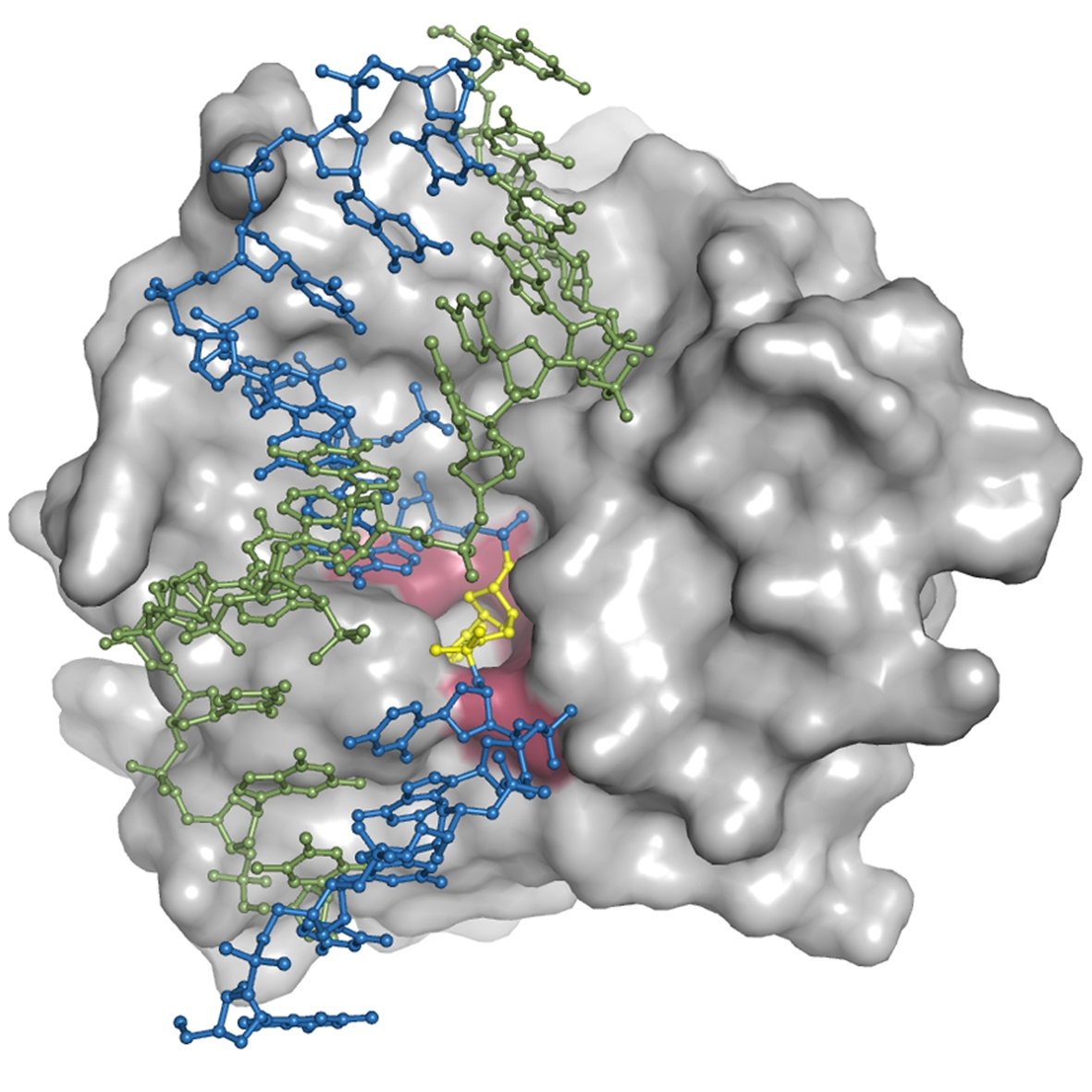
The DNA repair enzyme Endonuclease V can initiate the repair of deaminated purine bases by recognizing them and hydrolyzing the second phosphodiester bond on their 3' side. Deamination of DNA bases, particularly via nitrosative deamination resulting from endogenous processes and increased by oxidative stress from mitochondrial dysfunction or inflammatory responses, can cause transition mutations and cancer predisposition. Particularly, adenine (A) in DNA is transformed into the mutagenic hypoxanthine (inosine), which forms base pair with cytosine (C) instead of thymine (T), with a resulting A/T to G/C mutation upon replication.
In this project, we solved structures of endonuclease V in complex with its substrate and product DNA. These structures provide new insight into the initial step of what has been a structurally undefined, but biologically crucial, DNA base repair pathway The structures reveal a wedge motif acting as a minor groove damage sensor and a pocket to recognize the lesion; the enzyme remains tightly bound to the incised product, probably to hand it over to downstream repair proteins which are responsible for removal of the DNA damage and reinsertion of correct bases to restore the integrity of the DNA. The model further provedes a platform for analysis of genetic variations in the EndoV gene itself, particularly in the context of predispositions to cancer development.
Links:
Dalhus B, Arvai AS, Rosnes I, Olsen ØE, Backe PH, Alseth I, Gao H, Cao W, Tainer JA, Bjørås M (2009)
Structures of endonuclease V with DNA reveal initiation of deaminated adenine repair
Nat Struct Mol Biol (in press) (online 11 Jan 2009)
The web page of Magnar Bjørås' group - Laboratory for molecular biology
