Miriam Ragle Aure first author on article chosen as "editor's pick" in this month's Genome Biology. Defends thesis Nov 27.

Miriam Ragle Aure from Vessela Kristensen's Cancer Genome Variation group at Department of Genetics is first author on an article recently published in Genome Biology (journal impact factor 10.3). The article - entitled "Individual and combined effects of DNA methylation and copy number alterations on miRNA expression in breast tumors" - has been chosen as editor's pick" this month. The study evaluates the global effect of copy number and epigenetic alterations on miRNA expression in cancer.
Miriam Ragle Aure will defend her thesis entitled "From DNA to RNA to protein: integrated analyses of high-throughput molecular data from primary breast carcinomas" on Wednesday, November 27th at 13.15 in the Auditorium at IKF. Her trial lecture entitled "Computational and other approaches to distinguish between driver and passenger mutations of breast cancer" will be held at 10.15 the same day.
Summary of the findings, by the authors:
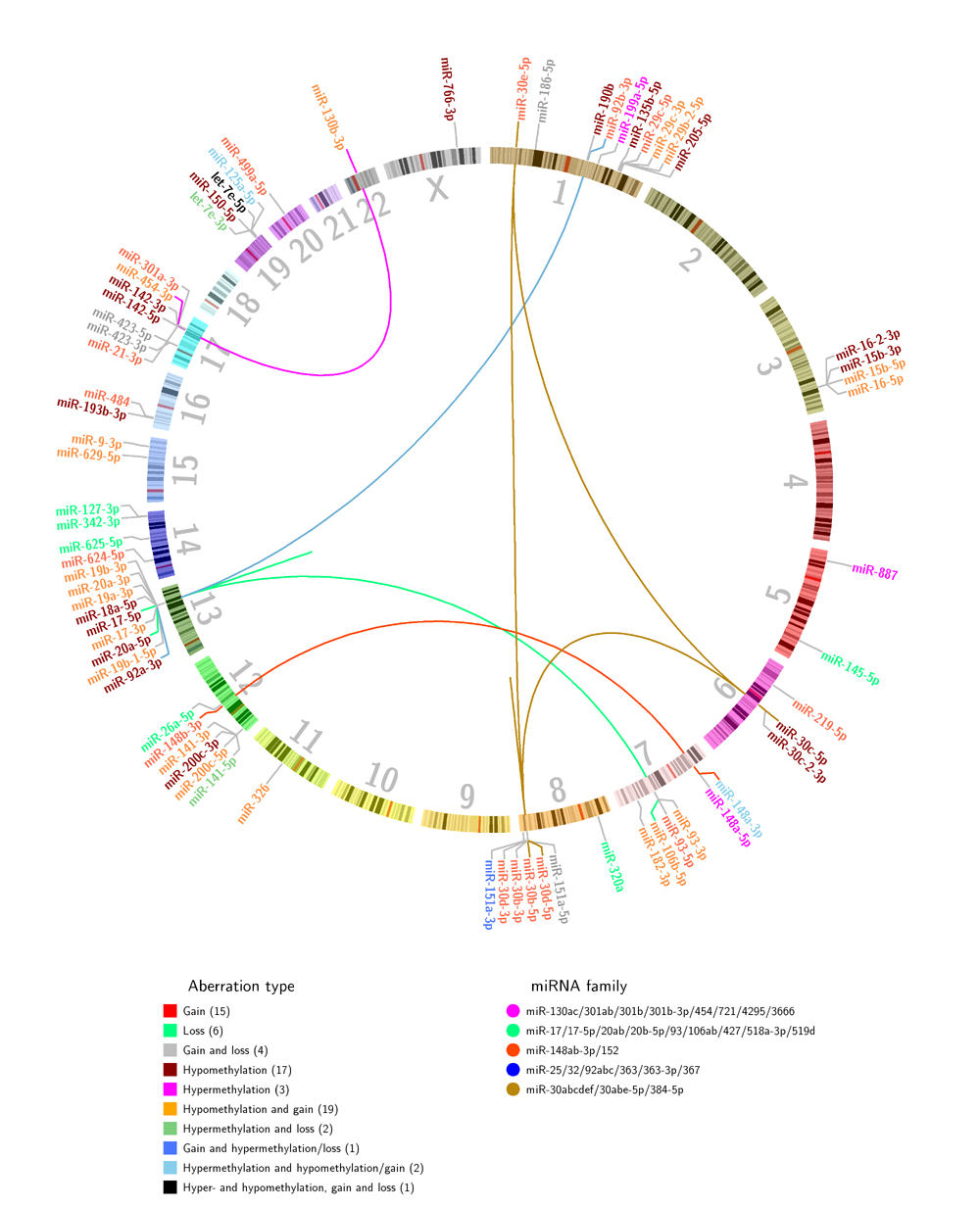
Genome-wide DNA methylation, copy number and miRNA expression was integrated to identify genetic mechanisms underlying miRNA dysregulation in breast cancer. 70 miRNAs whose expression was associated with alterations in copy number or methylation, or both were identified. Among these, five miRNA families were represented. Interestingly, the members of these families are encoded on different chromosomes and are complementarily altered by gain or hypomethylation across the patients. In an independent breast cancer cohort, 41 of the 70 miRNAs were confirmed with respect to aberration pattern and association to expression. In vitro functional experiments were performed in breast cancer cell lines with miRNA mimics to evaluate the phenotype of the replicated miRNAs. let-7e-3p, which in tumors was found associated with hypermethylation, was shown to induce apoptosis and reduce cell viability, and low let-7e-3p expression was associated with poorer prognosis. The overexpression of three other miRNAs associated with copy number gain, miR-21-3p, miR-148b-3p and miR-151a-5p, increased proliferation of breast cancer cell lines. In addition, miR-151a-5p enhances the levels of phosphorylated AKT protein. The data provide novel evidence of the mechanisms behind miRNA dysregulation in breast cancer. The study contributes to the understanding of how methylation and copy number alterations influence miRNA expression, emphasizing miRNA functionality through redundant encoding, and suggests novel miRNAs important in breast cancer.
Miriam R. Aure will defend her thesis entitled "From DNA to RNA to protein: integrated analyses of high-throughput molecular data from primary breast carcinomas" on Wednesday, November 27th at 13.15 in the Auditorium at IKF.
Her trial lecture entitled "Computational and other approaches to distinguish between driver and passenger mutations of breast cancer" will be held at 10.15 the same day.
Links:
Reference:
Aure MR, Leivonen SK, Fleischer T, Zhu Q, Overgaard J, Alsner J, Tramm T, Louhimo R, Alnæs GI, Perälä M, Busato F, Touleimat N, Tost J, Børresen-Dale AL, Hautaniemi S, Troyanskaya OG, Lingjærde OC, Sahlberg KK, Kristensen VN.
Individual and combined effects of DNA methylation and copy number alterations on miRNA expression in breast tumors. Genome Biol. 2013 Nov 20;14(11):R126. [Epub ahead of print]
Miriam Ragle Aure's disputation:
Details about the disputation from UiO's home page (in Norwegian)
Miriam Ragle Aure's publications
Home page of Vessela Kristensen's group - Cancer Genome Variation
Figure 1 from the article (click to enlarge):