Personalized medicine in clinical practice - is it feasible?
In a paper in BMC Cancer published October 20th - with Geir Olav Hjortland and Eivind Hovig as first and last author - a pilot case of trying to identify molecular tumor alterations as potential targets for therapy is demonstrated. The work represents a cross-disciplinary collaboration involving doctors at the Department of Oncology, the Department of Pathology, the Department of Medical Imaging and Intervention, and experimentalists from the Department of Tumor Biology at the Institute of Cancer Research. In cancer diagnostics today, such cross-disciplinarity is the norm. However, the hope is that this integration with the experimentalists will develop further, with the advent of molecular screening techniques.
Summary of the findings in the article "Genome wide single cell analysis of chemotherapy resistant metastatic cells in a case of gastroesophageal adenocarcinoma" by the authors:
When cancer hits a patient, this is a consequence of genetic changes, or mutations, in the cells of the patient. The genetic changes in a given tumor may vary in number and type between both tumor types and individual tumors. Commonly used cancer therapies, such as chemotherapy and radiation therapy, were originally developed to exploit general features of tumor, and were not tailored for the specific molecular changes in each tumor. The idea of analyzing each individual tumor in a cancer patient to find the best drugs to target the tumor of that particular patient, is an exiting new possibility that is now gradually becoming feasible. Extensive testing of the patient's tumor may reveal new potential targets for cancer therapy, and may in the future provide predictive information of cancer therapy options, often referred as "personalized medicine".
Screening for genetic changes in tumor cells is possible
Recently developed technologies such as massive parallel genomic sequencing now opens up for the identification of all possible genetic aberrations in tumor cells and to combine this information with the known targets of an increasing list of biology-based drugs being developed. Also, other whole genome scale analyses may be utilized to screen for potential drug targets for the tumor of a cancer patient. In a paper in BMC Cancer by Hjortland et al. published today, a pilot case of trying to screen for specific molecular tumor alterations as potential targets for therapy utilizing a molecular technique, called array comparative genomic hybridization, array CGH, to detect gene aberrations (i.e. gains or losses of genetic material) in tumors, enabling identification of gene alterations that may be targeted with drugs.
Novel therapies may be available for the cancer patient
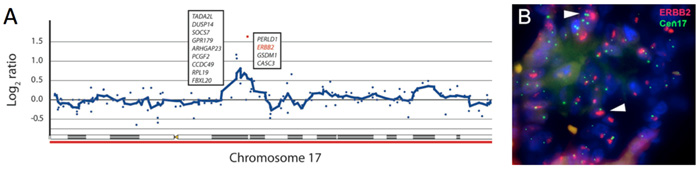
By a collaborative effort with several experimentalist colleagues fourteen cells from a bone metastasis spreading of metastatic gastroesophageal adenocarcinoma (a cancer of the digestive tract) were isolated. These 14 cells contained enough DNA for a successful analysis of the whole genome array CGH to allow identification of genetic changes, and it was possible to detect amplification of a gene called ERBB2 gene in these tumor cells (Figure A). The drug trastuzumab was developed to target of ERBB2 in breast cancer. Recently, trastuzumab has also shown efficacy in the treatment of metastatic gastric cancer, but this was unknown at the time of this case study.
The molecular targets could be used to monitor treatment response
Using molecular histopathology techniques, fluorescence in situ hybridizations (Figure B) and immunostaining, the findings from the genomic screening could be confirmed. However, it was also evident that the metastases in the bone did not respond to the standard treatment, and that the relative number of ERBB2 amplified cells in the bone metastasis had increased during the course of treatment.
The era of personalized medicine in cancer treatment is moving closer!
In this way, genome-wide DNA analysis of even very few tumor cells could be performed in a clinical setting during therapy, and the results suggested alternative targeted therapy strategies, as well as offering possibilities for monitoring of treatment response. Although this case illustrates the possibility of prospective global genomic testing of tumor cells, significant challenges remain when designing new clinical studies based on such extensive testing of tumor material. The results of genomic testing will need to be translated into predictive information regarding tumor directed therapy. In line with this, utilization of novel sequencing techniques to obtain even more complete characterization of tumors is already in progress at the Radium Hospital in a pilot study of lung cancer patient material, and efforts are underway to try to mobilize these techniques towards use in the cancer clinic.
For further information, contact Geir Olav Hjortland, email: Geir.Olav.Hjortland@rr-research.no
Links:
Geir Olav Hjortland, Leonardo A Meza-Zepeda, Klaus Beiske, Anne H Ree, Siri Tveito, Hanne Høifødt, Per J Bøhler, Knut H Hole, Ola Myklebost, Øystein Fodstad, Sigbjørn Smeland and Eivind Hovig:
Genome wide single cell analysis of chemotherapy resistant metastatic cells in a case of gastroesophageal adenocarcinoma. BMC Cancer 2011, 11:455. doi:10.1186/1471-2407-11-455 PubMed
The article is available at http://www.biomedcentral.com/1471-2407/11/455
|
|
| From left: Eivind Hovig (last author), Øystein Fodstad (scientific direktor at the Institute for Cancer Research and co-author of the article) and Geir Olav Hjortland (first author). |