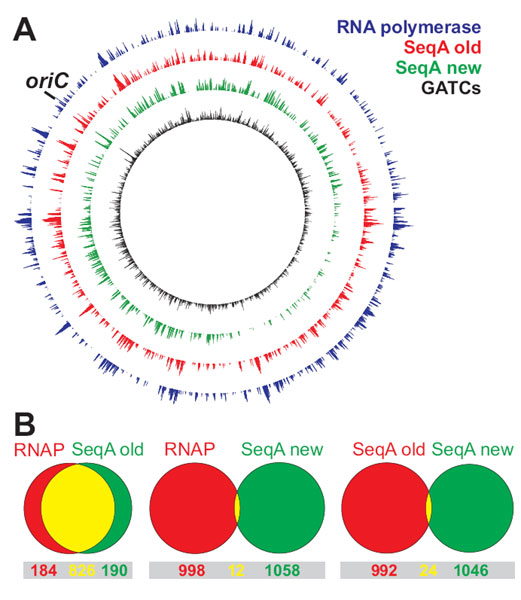
ChIP on Chip is a powerful method to detect genome wide binding of a protein (see box). Surprised by their own unlikely finding that the sequestration protein SeqA in E. coli has a binding pattern similar to the RNA polymerase (Fig. 1), Torsten Waldminghaus and Kirsten Skarstad started to investigate a much used ChIP on Chip protocol. Four causes of high background were discovered and the protocol modified accordingly. The new protocol gave very different results (Fig. 1). Reproduction of a study published by others revealed a false positive rate of about 50%. After publication of the results a lively discussion with researchers around the world has been going on.
Reference:
Waldminghaus T, Skarstad K. 2010. ChIP on Chip: surprising results are often artifacts.
BMC Genomics 5;11(1):414.
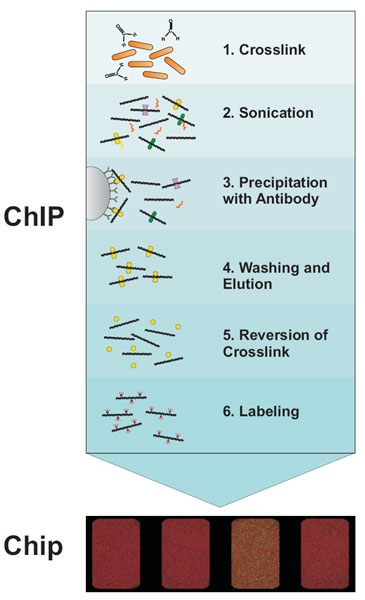
Box: How ChIP on Chip works
 |
 |
| Fig 1 |
Fig 2 |
|
The first step in the ChIP-Chip procedure is to fix protein-DNA interactions in living cells by chemical crosslinking with formaldehyde. After cell lysis the DNA is fragmented by sonication. This extract is then subjected to immunoprecipitation (IP) with a specific antibody against the protein of interest. DNA bound by the protein will be coprecipitated and enriched compared to DNA not bound by the respective protein. To facilitate immunoprecipitation and subsequent washing, antibodies are usually coupled to either agarose- or magnetic beads via protein A or G. After reversion of crosslinking the DNA is purified by phenol extraction or commercial PCR cleanup kits. Often, an amplification step is included after DNA purification. Two different fluorescence labels are used to label the IP DNA and a hybridization control DNA, respectively. Usually total DNA before IP (input DNA) is used as hybridization control. The two differentially labeled DNAs are hybridized to the same microarray and the difference in fluorescence intensity gives a measure of the enrichment.
|
ChIP on Chip signal on E. coli chromosome.
A First experiments gave a very similar signal of RNA polymerase (blue) and SeqA (red) binding. After modification of the method the SeqA signal (green) correlated well with the distribution of predicted binding sites (black).
B Venn diagrams show the overlap of ~1000 strongest ChIP on Chip signals of RNA polymerase (RNAP) and SeqA (see A). The modified and unmodified method give completely different results.
(click to enlarge image)
|
Links:
Torstein Waldminghaus
Home page of Kirsten Skarstad's group - Regulation of DNA replication
Department of Cell Biology
Institute for Cancer Research



