Long non-coding RNAs, circulating and disseminated tumor cells, and genome instability
Molecular signatures of long non-coding RNAs in breast cancer patients
Of the ~3.3 billion bases of the human genome, only about 2% code for proteins and until very recently, the remaining 98% have been considered to be 'junk'. However, large transcriptomic studies have shown that around 90% of the genome is actively transcribed of which a significant fraction may contribute to a previously underestimated layer of regulatory long non-coding RNAs (lncRNAs). In several studies it has been shown that the expression of small ncRNAs, like microRNAs, is associated with diseases including cancer. However, the large group of long lncRNAs has drawn less attention. LncRNA expression of selected 26 breast carcinoma samples, representing the five clinically relevant tumor expression subclasses, and 5 samples from normal breast tissue were analyzed using the nONCOchip®. The nONCOchip®, developed by the RNomics group at the Fraunhofer Institute for Cell Therapy and Immunology, covers both, experimentally identified cancer related lncRNAs as well as known or predicted non-coding RNAs from public databases. The array includes in total 243,000 probes, with over 60,000 newly identified transcripts. First results reveal that lncRNAs are variably expressed in breast tissue, expression signatures are different between breast tumors and normal breast tissues, expression patterns in breast cancer show a high degree of heterogeneity, and certain lncRNAs are significantly differentially expressed, e.g. between TP53 wild type and mutated samples. The overall aims of this study are to identify lncRNAs involved in gene regulation in breast cancer, to explore lncRNAs gene aberrations/expression profiles important for breast cancer development and progression, and to identify lncRNAs biomarkers predictive for breast cancer. As the growing list of lncRNA genes influencing carcinogenesis is striking, the identification of clinically relevant lncRNAs may open up for the development of novel biomarkers and therapeutic tools that attack diseased cells.
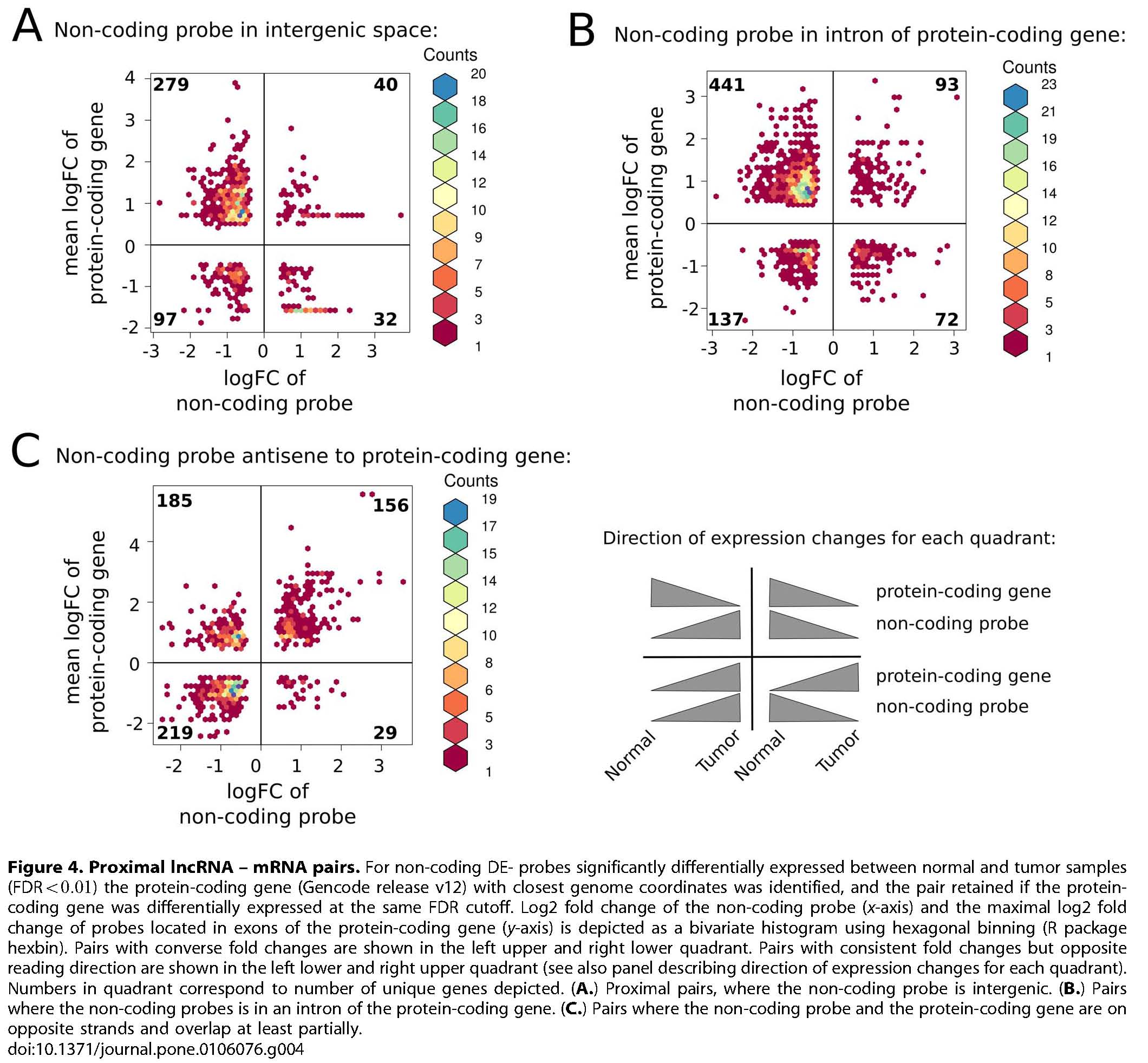
Reiche et al. (2014) PLoS ONE. DOI 10.1371/journal.pone.0106076, PubMed 25264628
This project resulted in the following publication:
Reiche K, Kasack K, Schreiber S, Lüders T, Due EU, B Naume, Riis M, Kristensen VN, Horn F, Børresen-Dale A-L, Hackermüller J, and Baumbusch LO (2014) Long non-coding RNAs differentially expressed between normal versus primary breast tumor tissues disclose converse changes to breast cancer-related protein-coding genes. PLoS ONE 9(9):e106076. doi.org/10.1371/journal.pone.0106076.
Molecular signatures of disseminated tumor cells in breast cancer
The main focus of this project group was to explore the genomic characteristics of disseminated tumor cells in breast cancer and to allocate their relevance to clinical parameters. The critical step in breast cancer progression is the establishment of metastasis to distant organs. Circulating tumor cells (CTC) in peripheral blood and disseminated tumor cells (DTC) in secondary organs like bone marrow are considered to be rare members among the cellular population of primary tumor cells. In several studies it has been demonstrated that the presence of these cells is an independent prognostic factor and detection of DTC identifies patients with less favorable clinical outcome. However, we still lack knowledge of the genomic characteristics of DTC and how to target these cells. A reason for this limitation has been a deficiency of procedures allowing high-resolution analyses of CTC/DTC, including DNA-copy number changes. Due to the implementation of a state-of-the-art technique, called single cell array comparative genomic hybridization (SCaCGH), we are finally able to perform in-depth analysis of occult circulating tumor cells on various high density array platforms. Further, new tools for detection and characterization of CTC/DTC including qRT-PCR have been developed. Genotyping and phenotyping of occult tumor cells will provide information to better understand tumor initiation, tumor heterogeneity, and subsequent metastasis formation. Using novel bioinformatic and biostatistical tools studied in connection with a detailed primary tumor analysis, we wish to identify biological markers and genes responsible for the capacity of cancer cells to metastasize. This may be clinically useful as evidence for an early occult spread of tumor cells, as a relevant prognostic factor, and finally, permit direct exploration of markers for targeted treatment. The project was supported by DISMAL. The main objective of DISMAL was to improve specificity and sensitivity of current platforms for DTC (disseminated tumor cells) detection in patients with epithelial tumors. The Goal was to identify novel markers at the DNA, RNA or protein level that allows a more precise detection of DTC with a high risk for metastatic progression. The project was an integrative part of the Breast Cancer study in the Micrometastasis project ongoing at the Norwegian Radium Hospital. The work was performed in close collaboration between the Dep. of Pathology and the Dep. of Genetics.
This project resulted in the following publications:
Demeulemeester J*, Kumar P*, Møller EK*, Nord S, Wedge DC, Peterson A, Mathiesen RR, Fjelldal R, Esteki MZ, Theunis K, Gallardo EF, Grundstad JA, Borgen E, Baumbusch LO, Børresen-Dale A-L, White KP‡, Kristensen VN‡, van Loo P‡, Voet T‡, and Naume B‡ (2017) Tracing the origin of disseminated tumor cells in breast cancer using single-cell sequencing. Genome Biology 17(1):250. doi.org/10.1186/s13059-016-1109-7.
Møller EK, Kumar P, voet T, Peterson A, van Loo P, Mathiesen RR, Fjelldal R, Grundstad J, Borgen E, Baumbusch LO, Naume B, Børresen-Dale A-L, White KP, Nord S, and Kristensen VN (2013) Next generation sequencing of disseminated tumor cells. Frontiers in Molecular and Cellular Oncology 3:320. doi.org/10.3389/fonc.2013.00320.
Otto B, Gruner K, Heinlein C, Wegwitz F, Nollau P, Ylstra B, Pantel K, Schumacher U, Baumbusch LO, Martin-Subero JI, Siebert R, Wagener C, Streichert T, Deppert W, and Tolstonog GV (2013) Low-grade and high-grade mammary carcinomas in WAP-T transgenic mice are independent entities distinguished by Met expression. International Journal of Cancer 132(6):1300. doi.org/10.1002/ijc.27783.
Mathiesen RR, Fjelldal R, Due EU, Geigl JB, Riethdorf S, Knut Liestøl, Borgen E, Rye IH, Schneider IJ, Obenauf AC, Mauermann O, Nilsen G, Lingjærde OC, Børresen-Dale A-L, Pantel K, Speicher MR, Naume B, and Baumbusch LO (2012) High resolution analysis of copy number changes in disseminated tumor cells of patients with breast cancer. International Journal of Cancer 131(4):E405-15. doi.org/10.1002/ijc.26444.
Molloy TJ, Bosma AJ, Baumbusch LO, Synnestvedt M, Borgen E, Russnes HG, Schlichting E, van't Veer L, and Naume B (2011) The prognostic significance of tumor cell detection in the peripheral blood versus the bone marrow in 774 early-stage breast cancer patients. Breast Cancer Research 13(3):R61. doi.org/10.1186/bcr2898.
Geigl JB, Obenauf AC, Waldispuehl-Geigl J, Fischer M, Trajanoski Z, Baumbusch LO, Speicher MR (2009) Identification of small gains and losses in single cells after whole genome amplification on tiling oligo arrays. Nucleic Acids Research 37(15):e105. doi.org/10.1093/nar/gkp526.
Genomic instability
Genomic instability is a hallmark of malignant tumours, causing disturbed integrity of the genome, numerical alterations, and structural changes. For various cancer types genomic instability has been associated with poor prognosis, suggesting that genomic instability may confer growth advantage of cancer cells. In most cancer types, genomic instability is characterized by copy number alterations, allelic imbalance, or the loss of heterozygosity. The molecular basis of genomic instability remains still unclear; however, mutations in key checkpoint proteins and DNA repair genes are supposed to be involved. Interestingly, the effects of disordered genomic organization may also have an unfavourable effect on the overall viability and fitness of cancer cells. Precise delineation of the negative and positive effects of genomic instability on cancer cells is of potentially great importance for tumour classification, survival prediction, and individualized therapy. The research is focused on the assessment of genomic instability in a variety of tumour types by applying various methods or the developing novel algorithms.This work includes projects about the complexity and genomic aberrations in breast tumors and serous ovarian cancers; integrated omics analysis; efficient algorithms for single- and multi-track copy number segmentation analysis; genomic architecture; comparison of platforms and algorithms for classification of copy number alterations in human breast tumors.
This project resulted in the following publications:
Kaveh F*, Baumbusch LO*, Nebdal D, Børresen-Dale A-L, Kingjærde OC, Edvardsen H, Kristensen VN, and Solvang HK (2018) Correction to: A systematic comparison of copy number alterations in four types of female cancer. BMC Cancer 18(1):80. doi.org/10.1186/s12885-017-3766-7.
Kaveh F*, Baumbusch LO*, Nebdal D, Børresen-Dale A-L, Kingjærde OC, Edvardsen H, Kristensen VN, and Solvang HK (2016) A systematic comparison of copy number alterations in four types of female cancer. BMC Cancer 16(1):913. doi.org/10.1186/s12885-016-2899-4.
Liu Y*, Zhou R*, Baumbusch LO, Tsavachidis S, Brewster AM, Do K-A, Sahin A, Hortobagyi GN, Taube JH, Mani SA, Aarøe J, Wärnberg F, Børresen-Dale A-L, Mills GB, Thompson PA, and Bondy ML (2014) Genomic copy number imbalances associated with bone and non-bone metastasis of early-stage breast cancer. Breast Cancer Research and Treatment 143(1)189. doi.org/10.1007/s10549-013-2796-3.
Baumbusch LO*, Helland Å*, Wang Y, Liestøl K, Schaner ME, Holm R, Etemadmoghadam D, Alsop K, Brown P, AOCS Group, Mitchell G, Fereday S, Defazio A, Bowtell DDL, Kristensen GB, Lingjærde OC, and Børresen-Dale A-L (2013) High levels of genomic aberrations in serous ovarian cancers are associated with better survival. PLoS ONE 8(1):e54356. doi.org/10.1371/journal.pone.0054356.
Aure MR*, Steinfeld I*, Baumbusch LO*, Liestøl K, Lipson DK, Naume B, Kleivi K, Kristensen VN, Børresen-Dale AL, Lingjærde OC, and Yakhini Z (2013) Identifying in-trans process associated genes in breast cancer by integrated analysis of copy number and expression data. PLoS ONE 8(1):e53014. doi.org/10.1371/journal.pone.0053014.
Nilsen G, Listøl K, van Loo P, Vollan HKM, Eide MB, Rueda OM, Chin S-F, Russell R, Baumbusch LO, Caldas C, Børresen-Dale A-L, and Lingjærede OC (2012) Copynumber: Efficient algorithms for single- and multi-track copy number segmentation analysis. BMC Genomics 13:591. doi.org/10.1186/1471-2164-13-591.
Russnes HG*, Vollan HKM*, Lingjærde OC, Krasnitz A, Lundin P, Naume B, Sørlie T, Borgen E, Rye IH, Langerød A, Chin S-F, Teschendorff AE, Stevens PJ, Månér S, Schlichting E, Baumbusch LO, Kåresen R, Stratton MP, Wigler M, Caldas C, Zetterberg A, Hicks J, and Børresen-Dale A-L (2010) Genomic architecture characterizes tumor progression paths and fate in breast cancer patients. Translational Science 2(38):38ra47. doi.org/10.1126/scitranslmed.3000611.
(The marked authors should be considered as joint first (*) or senior (‡) authors)
