Prostate Cancer
Prostate cancer is the second most common cause of cancer death in men in the developed world. Since the 1990s, early detection methods, such as screening for PSA (prostate-specific antigen) have been used to stratify patients at risk of death due to prostate cancer.
Whilst screening for prostate cancer can detect the disease early, it is also controversial. The standard screening process is unable to determine how aggressive the tumour is going to be, or what type and degree of treatment is needed. This means that men diagnosed with prostate cancer may end up receiving far more treatment, and further invasive testing, than is really needed for their cancer; all of which can have a very negative effect on the patient’s quality of life.

Responsive DNA accessibility to epigenetic drug
Accessibility of DNA varies from patient to patient, and may act as a fingerprint for prostate cancer progression.
Studying the specific characteristics of such DNA accessibility could make it possible to understand which of the epigenetics treatments already used in clinical trials for other cancers are best suited to prostate cancer patients.
Precision medicine and cancer treatment for prostate cancer patients
Another challenging promise of “precision medicine” in cancer is to establish differences between tumours. These differences can be used to differently target defects of the cells that have allowed the cancer to grow and escape conventional chemotherapy. Ultimately, this would mean scheduling patients for different treatments depending on the different defects that have allowed the tumour to grow.
To understand such so-called biomarkers for treatment is, essentially, trivial but it does hold the key to a future where we can better use drugs that are already available, through re-purposing them for treating other diseases, such as those that are apparently different but genetically similar.
Androgen Receptor and prostate cancer
The Androgen Receptor (AR) is a protein that fuels prostate cancer growth, by ‘switching on’ genes that wouldn’t normally be associated with the disease. Androgens, or male sex hormones such as testosterone, send messages through the AR, into the prostate’s cells and tissues. If there is a fault in this messaging, cell division can be triggered which can lead to abnormal growth of the cells and, therefore, prostate cancer.
Standard treatments for prostate cancer involve targeting and blocking the AR, however some men can become immune to this therapy. This is when castration-resistant prostate cancer (CRPC) can develop, for which many drugs have been approved but, despite prolonged survival, none of them can ultimately ‘cure’ the patient.
Chromatin relaxation
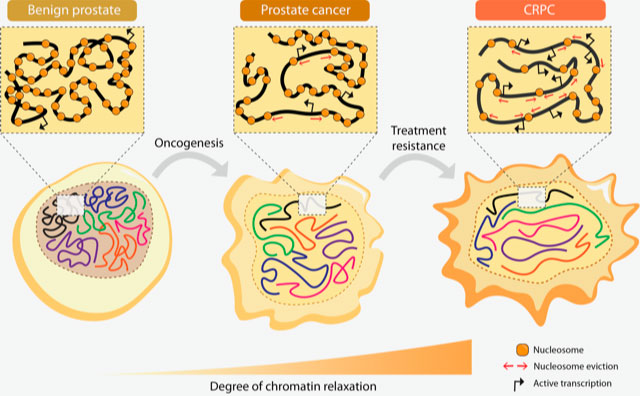
We have shown how upregulated AR and bromodomain proteins regulated by AR, contribute to genome-wide chromatin relaxation, which then leads to increased DNA accessibility. This in turn leads to increased gene transcription in advanced prostate cancer. The study showed, in very basic terms, that the relaxed chromatin is a typical feature of advanced prostate cancer.
Bromodomain-containing proteins (BRDs), such as BRD4, and androgen-regulated BRD2 and ATAD2, are the mediators of such increased DNA accessibility, and can be prognostic tissue markers. For instance ATAD2 was strikingly present in the tumours of those patients that relapsed after first treatment via androgen deprivation therapy.
New prognostic and predictive tools for prostate cancer
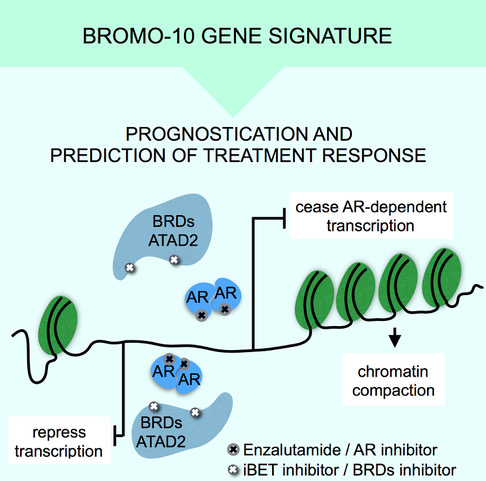
These findings constitute the potential for creating new prognostic and predictive tools, whereby chromatin accessibility can be used as an indicator of prostate cancer progression and severity, and help indicate the most suitable form of treatment for the patient.
By studying the chromatin and DNA accessibility, we have also determined a 10-gene signature, BROMO-10, that can be used as a genetic biomarker to show which patients fall into different categories in terms of the severity of their cancer.
Using BROMO-10, they also predicted which tumours responded to treatments with bromodomain inhibitors, thereby helping to provide further tools for personalising prostate cancer treatment.
Gene regulation and prostate cancer
We found that c-Myc and the AR regulate the expression of important genes for prostate cancer progression. Importantly, many genes are commonly regulated by both c-Myc and AR. The key finding of this study is that c-Myc, which was generally thought to be a generalist amplifier of transcription, is instead able to antagonise the expression of some of these important genes, such as PSA. PSA is the well-known prostate cancer biomarker used in the clinic to diagnose and follow up prostate cancer patients.
c-MYC overexpression has been previously observed in prostate cancer. However, this is the first time that c-Myc has been shown to deregulate the transcriptional program promoted by the AR.
These findings provide important insights that may have implications for precision medicine. Namely, this study provides evidences for helping to determine why PSA or other biomarkers may fail to identify the right treatment for some patients.
The study was also highlighted in Nature Reviews Urology
Epigenetics and risk of developing prostate cancer
We also evaluated how a generic epigenetic factor, the epigenetic reader Bromodomain containing protein 4 (BRD4), is able to flag areas of DNA that vary within individuals, and that are associated with several diseases such as different types of cancers, but also with neuropsychiatric disorders.
In collaboration with the group of Prof. Ole Andreassen at the NORMENT school, we studied the capacity of BRD4 to annotate genetic variants that are of functional importance in these diseases.
Epigenetics and disease prediction
A patient’s risk of developing cancer can often be identified through the identification of DNA-sequence variants, such as single nucleotide polymorphisms (SNPs). SNPs can also help predict a patient’s response to a drug, or their susceptibility to environmental factors.
Interestingly, epigenetics – the study of heritable changes in gene activity that do not involve alterations to the genetic code – are proposed to be useful in helping to identify clinically relevant SNPs that are of functional importance in cancer development.
Super-enhancers are cell-specific DNA elements that act to determine tissue or cell identity and drive tumour progression. Although previous approaches have attempted to explain the risk associated with SNPs in regulatory DNA elements, so far epigenetic readers, such as BRD4 and super-enhancers have not been used to annotate SNPs. In prostate cancer, androgen receptor (AR) binding sites to chromatin, are so far the only read-out used to inform functional annotations of SNPs.
BRD4 key discriminant of tissue-specific enhancers
We demonstrated that BRD4 is the key discriminant of tissue-specific enhancers, and shown that BRD4 it is more powerful than other previously studied factors such as AR binding information to identify prostate cancer-specific risk loci.
Furthermore, we showed that this approach is applicable to other diseases and traits, such as breast cancer and schizophrenia.
Proteins responsible for reading epigenetic marks on gene expression are increasingly becoming a target for drug development. These proteins bind DNA areas, which function as regulatory elements for the expression of genes, similar to genetics switches. The sequence embedded in such genetic switches may vary from individual to individual. Some variation is associated with an increased risk of developing diseases such as cancer or, as shown by us, neuropsychiatric disorders.
This study also provides a powerful tool for prioritizing genetic risk DNA variants (SNPs) for functional valiudation.
