New publication from the lab with contribution from several international collaborators Single-cell ATAC and RNA sequencing reveal pre-existing and persistent cells associated with prostate cancer relapse
Significance
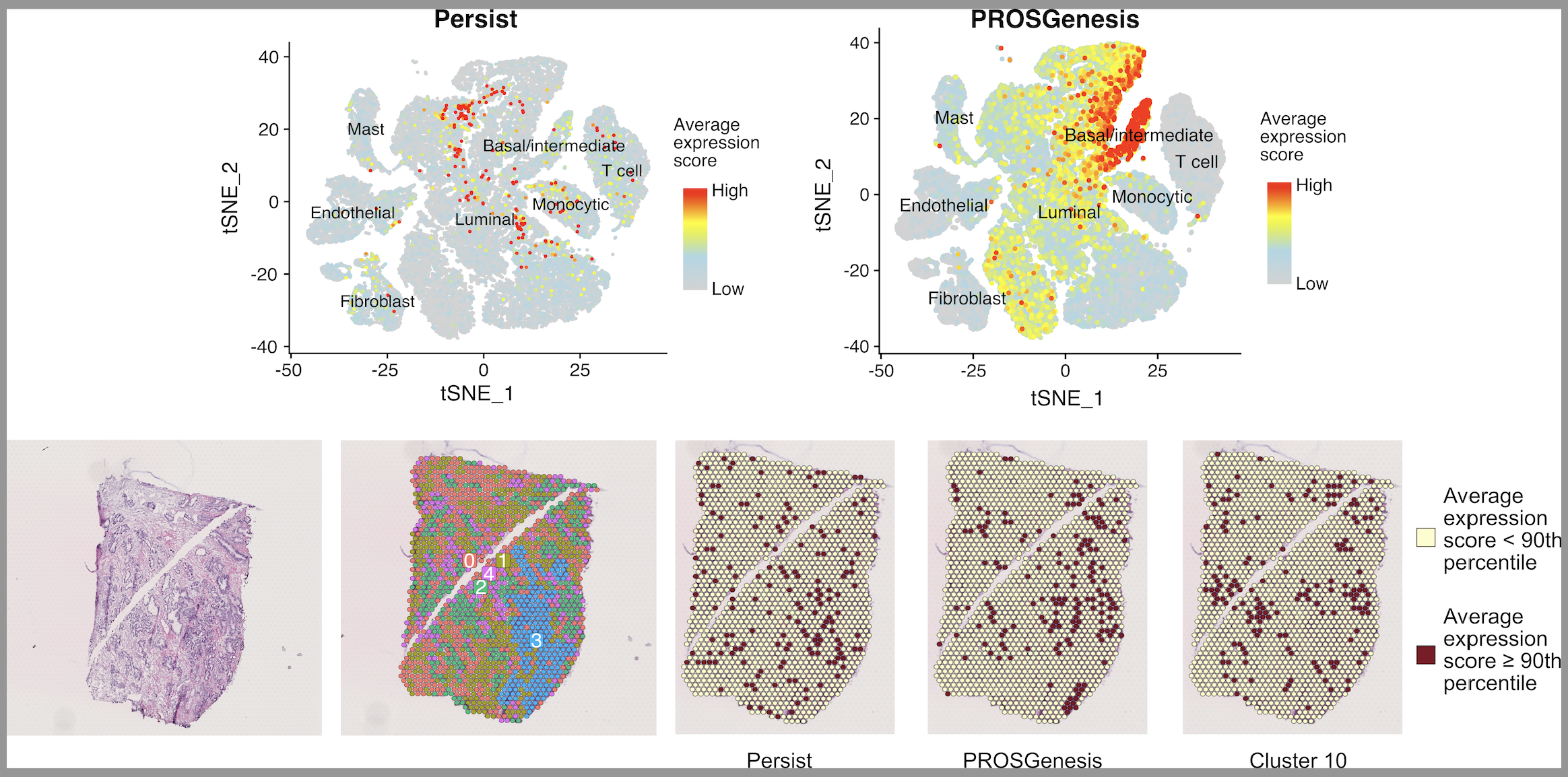
We used models of resistance to approved androgen receptor-targeted therapies for prostate cancer to identify subpopulations of treatment-persistent and pre-existing cells. We established that chromatin structure reconfigurations led to alterations in gene expression and drove alternative tumor adaptations and treatment escape. Motivated by the need for pre-treatment biomarkers in prostate cancer, we identified molecular predictors of therapy response based on the presence of treatment-persistent and pre-existing cells.
Summary
We show that subpopulations of treatment-persistent cells with stem-like and regenerative properties foster alternative trajectories of enzalutamide resistance in prostate cancer. Alternative transcriptional patterns of resistance are induced by divergent chromatin reprogramming. Transcriptional enrichment of signals from these treatment-persistent cells stratifies patient outcomes in both early stage treatment-naive and treatment-exposed tumors.
Highlights
- Identification of prostate cancer cells with gene expression patterns of regenerative potential that persist and exist prior to enzalutamide treatment.
- Profiling of chromatin and transcriptional features from subpopulations of treatment-challenged prostate cancer cells.
- Identification of gene signatures associated with stem-like and regenerative potential.
- Stratification of prostate cancer patients from “bulk” RNA sequencing data based on identified stemness- and regeneration-related gene signatures.
link to the study here
