Structural DNA and RNA variation
- including fusion genes, as a source of cancer biomarkers
This is a subproject of the Cancer Genomics project at the Department of Molecular Oncology.
Cancer cells commonly have structurally altered chromosomes (translocations, insertions, deletions, and duplications), and this may give rise to fusion genes producing chimeric RNA molecules. Chimeric RNA may also be caused by mechanisms such as polymerase read-through and trans-splicing. Certain chimeric RNAs are specific to cancer cells and are, as such, ideal as cancer biomarkers and drug targets.
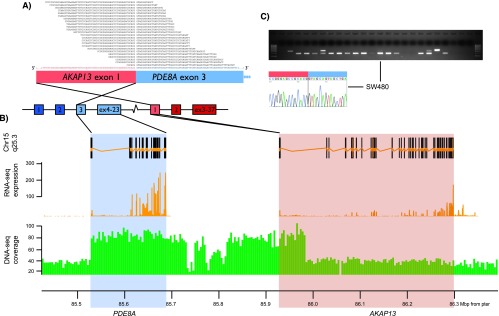
The research group uses both long- and short-read high-throughput sequencing of DNA and RNA to detect known and novel fusion genes in cancer. Several recurrent fusion transcripts have been discovered from testicular (Hoff et al., 2016), colorectal (Nome et al., 2013 and 2014), prostate (Zhao et al., 2017, Paulo et al., 2012, and Barros-Silva et al., 2013), and ovarian cancers (Smebye et al., 2017).
RNA variations are also arising due to alternative and aberrant pre-mRNA splicing. This phenomenon is covered by several papers by the group (Strømme et al., 2023; Hoff et al., 2016; Sveen et al., 2014; and Løvf et al., 2014) and has also been extensively reviewed (Sveen et al., 2016).
The group has also developed various research tools for the detection and study of structural DNA and RNA variants. The ScaR software is a bioinformatics tool for sensitive detection of fusion transcripts from RNA-seq data (Zhao et al., 2020), and chimeraviz is software for visualization of fusion genes (Lågstad et al., 2017). Transcriptome instability can be studied by the R package TIN (Johannessen et al., 2015). A novel oligonucleotide microarray-based system for fusion gene detection was developed (Skotheim et al., 2009 and Løvf et al., 2011).
Selected publications
Strømme JM, Johannessen B, and Skotheim RI (2023). Deviating alternative splicing as a molecular subtype of microsatellite stable colorectal cancer. JCO Clin Cancer Inform. 7: e2200159
Kidd SG*, Bogaard M*, Carm KT, Bakken AC, Maltau AMV, Løvf M, Lothe RA, Axcrona K, Axcrona U**, and Skotheim RI** (2022). In situ expression of ERG protein in the context of tumor heterogeneity identifies prostate cancer patients with inferior prognosis. Mol. Oncol. 16(15): 2810-22 Shared *first/**senior authors
Zhao S*, Hoff AM*, and Skotheim RI (2020). ScaR - A tool for sensitive detection of known fusion transcripts: Establishing prevalence of fusions in testicular germ cell tumours. NAR Genom. Bioinform. 2(1): 1-12 *Equal contribution
Lågstad S, Zhao S, Johannessen B, Hoff AM, Lingjærde OC, and Skotheim RI (2017). Chimeraviz: A Bioconductor package for visualizing chimeric RNA. Bioinformatics 33(18): 2954-6
Sveen A, Kilpinen S, Ruusulehto A, Lothe RA, and Skotheim RI (2016). Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene 35(19): 2413-27
Hoff AM, Alagaratnam S, Zhao S, Bruun J, Andrews PW, Lothe RA, and Skotheim RI (2016). Identification of novel fusion genes in testicular germ cell tumors. Cancer Res. 76(1): 108-16
Johannessen B, Sveen A, and Skotheim RI (2015). TIN: An R Package for Transcriptome Instability Analysis. Cancer Inform. 14: 109-12
Løvf M, Nome T, Bruun J, Eknaes M, Bakken AC, Mpindi JP, Kilpinen S, Rognum TO, Nesbakken A, Kallioniemi O, Lothe RA, and Skotheim RI (2014). A novel transcript, VNN1-AB, as a biomarker for colorectal cancer. Int. J. Cancer 135(9): 2077-84
Sveen A, Johannessen B, Teixeira MR, Lothe RA, and Skotheim RI (2014). Transcriptome instability as a molecular pan-cancer characteristic of carcinomas. BMC Genomics 15(1): 672
Nome T, Hoff AM, Bakken AC, Rognum TO, Nesbakken A, and Skotheim RI (2014). High frequency of fusion transcripts involving TCF7L2 in colorectal cancer: novel fusion partner and splice variants. PLoS One 9(3): e91264
Nome T, Thomassen GOS, Bruun J, Ahlquist TC, Bakken AC, Rognum T, Nesbakken A, Lorenz S, Sun J, Barros-Silva JD, Lind GE, Myklebost O, Teixeira MR, Meza-Zepeda LA, Lothe RA, and Skotheim RI (2013). Common fusion transcripts identified in colorectal cancer cell lines by high throughput RNA sequencing. Transl. Oncol. 6(5):546-53
Barros-Silva JD, Paulo P, Bakken AC, Cerveira N, Løvf M, Henrique R, Jerónimo C, Lothe RA, Skotheim RI, and Teixeira MR (2013). Novel 5' fusion partners of ETV1 and ETV4 in prostate cancer. Neoplasia 15(7): 720-726
Paulo P, Barros-Silva JD, Ribeiro FR, Ramalho-Carvalho J, Jerónimo C, Henrique R, Lind GE, Skotheim RI, Lothe RA, and Teixeira MR (2012). FLI1 is a novel ETS transcription factor involved in gene fusions in prostate cancer. Genes Chromosomes. Cancer 51(3): 240-249
Løvf M*, Thomassen GOS*, Bakken AC, Celestino R, Fioretos T, Lind GE, Lothe RA, and Skotheim RI (2011). Fusion gene microarray reveals cancer type-specificity among fusion genes. Genes Chromosomes. Cancer 50: 348-357 *Equal contribution
Skotheim RI, Thomassen GOS, Eken M, Lind GE, Micci F, Ribeiro FR, Cerveira N, Teixeira MR, Heim S, Rognes T, and Lothe RA (2009). A universal assay for detection of oncogenic fusion transcripts by oligo microarray analysis. Molecular Cancer 8(1): 5
