Welcome to Chloe B. Steen’s project group Cancer Biology in Silico

Project group leader
By leveraging different types of gene expression data, we've shown that employing a computational approach can overcome the limitations of experimental methods in the analysis of tissue and cancer heterogeneity. Our research aims to develop and apply computational methods for the identification of rare cell states and their respective microenvironments, with the goal of finding novel therapeutic targets and improving patient outcomes.
The project group is part of the Cancer Genome Variation group, led by Prof. Vessela Kristensen, with vast experience in breast cancer research and genomics.
In addition, Chloe has a 20% position in the Lymphoma Biology Group, led by June H. Myklebust at the Institute for Cancer Research (OUS), and is an associated member of the Newman lab at Stanford University.
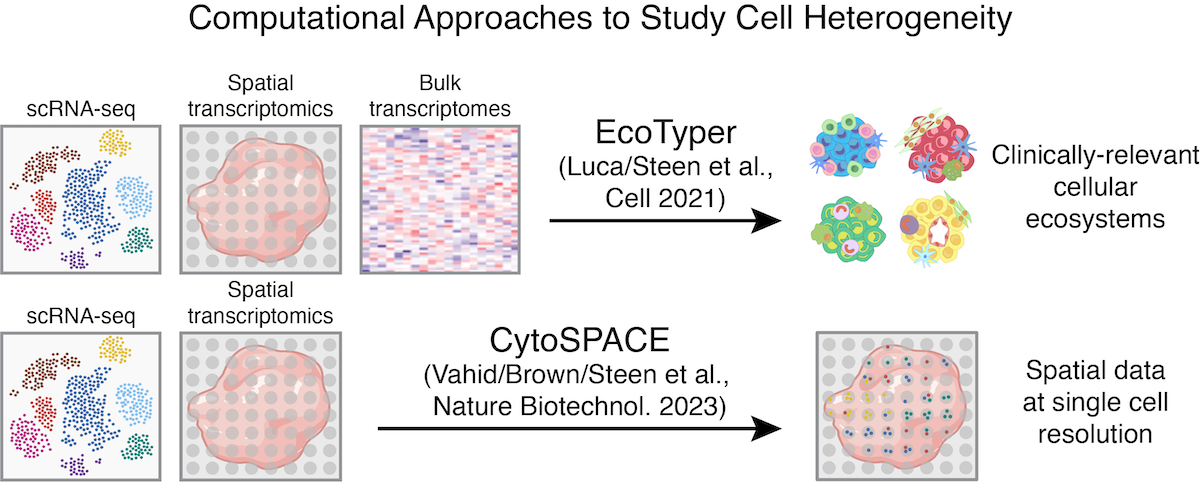
Key publications
- Vahid MR*, Brown EL*, Steen CB*, [...], Newman AM. High-resolution alignment of single-cell and spatial transcriptomes with CytoSPACE. Nature Biotechnology 2023. DOI 10.1038/s41587-023-
01697-9, PubMed 36879008 - Luca BA*, Steen CB*, [...], Gentles AJ, Newman AM. Atlas of clinically distinct cell states and ecosystems across human solid tumors. Cell 2021. DOI 10.1016/j.cell.2021.
09.014, PubMed 34597583 - Steen CB*, Luca BA*, [...], Newman AM, Alizadeh AA. The landscape of tumor cell states and ecosystems in diffuse large B cell lymphoma. Cancer Cell 2021. DOI 10.1016/j.ccell.
2021.08.011, PubMed 34597589 - Steen CB, [...], Newman AM. Profiling Cellular Ecosystems at Single-Cell Resolution and at Scale with EcoTyper. Methods Mol Bio. 2023 DOI 10.1007/978-1-0716-
2986-4_4, PubMed 36929073 - Steen CB, [...], Newman AM. Profiling Cell Type Abundance and Expression in Bulk Tissues with CIBERSORTx. Methods Mol Bio. 2020. DOI 10.1007/978-1-0716-
0301-7_7, PubMed 31960376
