FRIPRO Funding Awarded for Pioneering Research in Digital Pathology and Artificial Intelligence

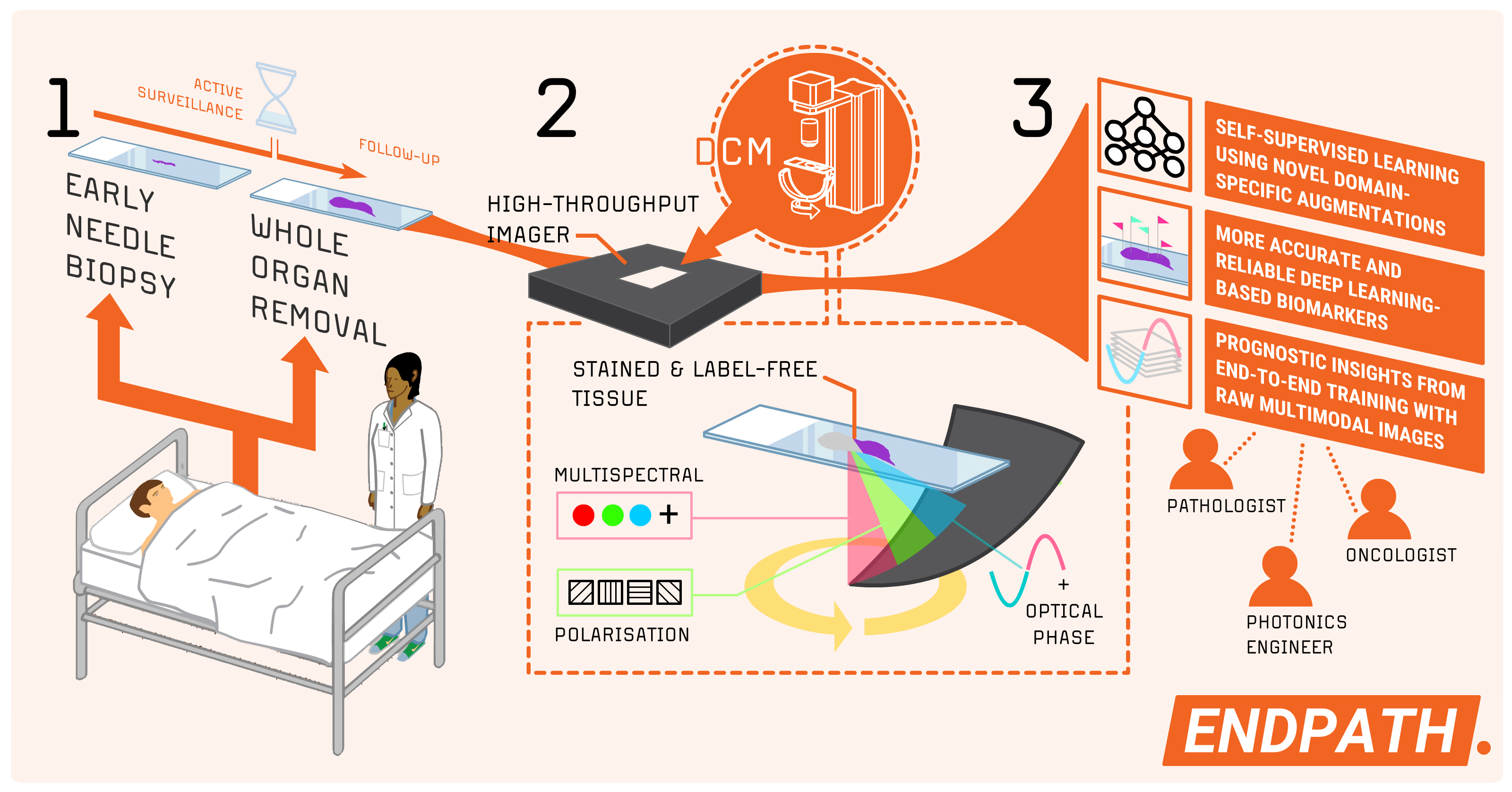
Research Director Andreas Kleppe, at the Institute for Cancer Genetics and Informatics (ICGI), has been awarded funding from the Norwegian Research Council's FRIPRO scheme for the ENDPATH – End-to-End Pathology project. This initiative sets out to develop a new imaging system for digital pathology, using it to train artificial intelligence (AI) models that could improve predictions of cancer progression. The project, initially focusing on prostate cancer, aims to enhance both the accuracy and reliability of diagnostic tools.

By moving beyond traditional imaging methods tailored for human interpretation, ENDPATH seeks to optimize image acquisition for AI-based analysis. This approach opens the door to opportunities for more accurate, reliable automatic pathology and deep learning with ground-breaking, multimodal, high-throughput computational imaging systems.
For more than 20 years, the ICGI has been at the forefront of improving cancer diagnostics by combining biomedicine and informatics. Led by researchers like Dr. Andreas Kleppe, the institute has played a key role in introducing AI and deep learning into tissue diagnostics. Through the DoMore project (2016–2021), also funded by the Norwegian Research Council (Lighthouse scheme), the ICGI helped advance the transition from digital pathology to in silico pathology, delivering significant innovations, including the DoMore-v1-CRC model. Their groundbreaking work, published in The Lancet, The Lancet Oncology, and The International Journal of Cancer, exemplifies the institute’s impact on advancing cancer prognostics and enabling personalized treatment strategies.
Guided treatment decisions for prostate cancer patients with DCM imagery
Prostate cancer is the most common cancer type among men in Norway, with significant potential benefits in avoiding unnecessary surgery. Currently, over 6,000 patients with prostate cancer are enrolled in active surveillance programs across the country, highlighting the importance of accurate risk assessment to guide treatment decisions.
One of the tasks in the ENDPATH project will be on extracting actionable prognostic insights through the first large-scale pathology study of its kind. Using a unique dataset of more than 2,000 prostate cancer samples, we aim to demonstrate the clinical advantages of DCM over conventional microscopy.
To optimize the use of this data, the project team will employ advanced self-supervised learning frameworks, specifically tuned for pathology tasks. The trained models will first address key tasks such as tumor segmentation and Gleason grading, allowing visual and pathological feedback to verify that the DCM images, vision transformers, and downstream task adaptations work as intended. Models for prognostic evaluations will then be adapted, providing valuable tools to improve decision-making in prostate cancer management.
The method of choice has the potential of making routine pathology imaging more robust. The project will therefore aim to establish image quality assurance protocols tailored for clinical use.
Expanding the Potential of Digital Pathology
For the last 150 years, hematoxylin-eosin (H&E) staining has been, and remains, the gold standard in pathology. Current imaging systems in the clinic primarily capture intensity and color, but leave other properties of light—such as phase, amplitude, and polarization—unutilized. We believe these properties hold significant potential when leveraged through deep learning.
Unlike slower and more complex imaging methods, DCM allows for fast, cost-effective image acquisition while preserving image appearance familiar to routine pathologists and compatible with the existing clinical decision frameworks. Building on recent advancements, Kleppe and his group constructed a DCM imager prototype based off a high-throughput microscopy framework. By continuing this work in “End-to-end Pathology”, the team aims to create the first multimodal DCM system for advanced deep learning. The system will be optimized for clinical pathology, but has a potential of technological ripple effects in other fields such as affordable microscopy and photonics.
Partners and collaboration
The ICGI is grateful for the opportunity to collaborate with renowned institutions such as the University of South-Eastern Norway (USN) and University College London (UCL). Building on ICGI’s strong track record in developing cutting-edge AI tools for cancer diagnostics and prognostics, these collaborations bring together expertise in imaging technology, computational modeling, and clinical applications, ensuring that the project benefits from diverse perspectives and insights. In addition, skilled colleagues at the Institute for Informatics at the University of Oslo, where Kleppe holds a position as Associate Professor, are eager to contribute.
An advisory board consisting of Dr. Andreas Kleppe, Dr. Mike Shaw, Prof. Muhammad Nadeem Akram, Dr. Hanne Askautrud, Dr. Ole-Johan Skrede, Ilya Kostolomov, Dr. Tarjei Hveem and Prof. Marco Novelli will oversee the project.
Fierce competition – high quality
FRIPRO is a national funding scheme for groundbreaking research known to be highly competitive. Only applications that receive top marks on all assessment criteria are eligible for funding. The fact that Andreas Kleppe and this project are among the selected few, is clear proof of both quality and originality. “At FRIPRO, we are willing to invest in the bold research that has the potential to make significant advances in the field, even if it also has a significant risk of failure,” the Norwegian Research Council states. The ICGI at Oslo University’s Cancer Clinic has ample experience within digital imaging and AI, and the research group is eager to explore new horizons and shape the next generation of digital pathology.
Links:
News article (in Norwegian):
FRIPRO-midler til banebrytende forskning på digitalpatologi og kunstig intelligens (KI) (oslo-universitetssykehus.no)
From the Norwegian Research Council - about this year's FRIPRO awards:
Toppforskere får 191 millioner kroner (forskningsradet.no)
About FRIPRO - Forskerprosjekt for tidlig karriere (forskningsradet.no)
