Nature Communications publication: Single-molecule experiments reveal how DNA repair enzymes look for damages using a combination of helical sliding and jumping

Researchers at Department for Microbiology, Department for Medical Biochemistry and Department for Newborn Screening have used single-molecule fluorescence microscopy to track single protein molecules as they move along a linear piece of DNA held in place by the use of a laser/optical tweezers. By tracking single protein molecules as they moved along the DNA molecule, the interaction between protein and DNA could be investigated in detail in real-time. This study has been performed by PhD student Arash Ahmadi and co-workers in the group of Bjørn Dalhus at the Department for Microbiology, OUS, and Department for Medical Biochemistry, UiO. The study is part of a collaboration between OUS, UiO, NTNU and the University of Bielefeld, Germany.
The DNA of cells are constantly exposed to internal and external factors that pose a constant threat to the integrity of this valuable molecule, which only exists in a single copy in non-dividing cells. A plethora of DNA repair proteins protects the DNA from damages that would otherwise give mutations, which could later lead to chromosomal instability, diseases or premature aging. It has been estimated that a single cell in a human body can experience tens of thousands of damages every day on average. Thus, efficient localization of damages in the DNA is essential for keeping DNA in good condition. While the process of DNA damage repair has been extensively studied for decades by molecular and cell biology, with major contributions from many Norwegian researcher, the dynamics and choreography of single DNA repair proteins operating on DNA is limited.
- In this study we have investigated how the DNA repair proteins Endonuclease V and Ogg1 scan DNA using different strategies to search for the damages among a vast majority of normal DNA bases, says Bjørn Dalhus. We have used super-resolution fluorescence microscopy to look at single protein molecules as they move along DNA in search for a potential damage to repair. To be able to investigate this process using live imaging with a resolution high enough to visualize single molecules, we used a laser/optical tweezers to trap and stretch a single DNA molecule to its full length and then added fluorescently labeled DNA repair proteins. It’s worth mentioning that the technology of optical tweezers was awarded the Nobel Prize in physics earlier this year. We could then record high-frequency movies of single DNA repair proteins scanning DNA in search for damages. By tracking single protein molecules as they moved along the linear DNA, we could study and characterize the interaction between protein and DNA in detail.
Analysis of the protein trajectories revealed that the Endonuclease V protein uses a mix of helical 'spnning' around the DNA double-helix and short jumps along the DNA to cover the DNA as efficiently as possible. The molecule stays close to the DNA also when it jumps from one site to the next. The two enzymes investigated in this study uses 'base flipping' to inspect single DNA bases for any damage, and we could also observe how the enzymes stopped along the DNA for a closer inspection of the DNA, says Dalhus. Finally, by mutating a motif in the Endonuclease V enzyme the research team could show that this motif is involved in DNA damage inspection while scanning the DNA.
The study, led by Bjørn Dalhus, Department for Medical Biochemistry and Department for Microbiology, and Alexander D Rowe, Department for Newborn Screening, is published in the prestigious journal Nature Communications, entitled "Breaking the speed limit with multimode fast scanning of DNA by Endonuclease V". The work has been performed in collaboration with Mark Schüttpelz and Robin Diekmann from the University of Bielefeld, Germany, Kyrre Glette and Jim Tørresen from Department of Informatics, UiO, and Magnar Bjørås from NTNU and OUS.
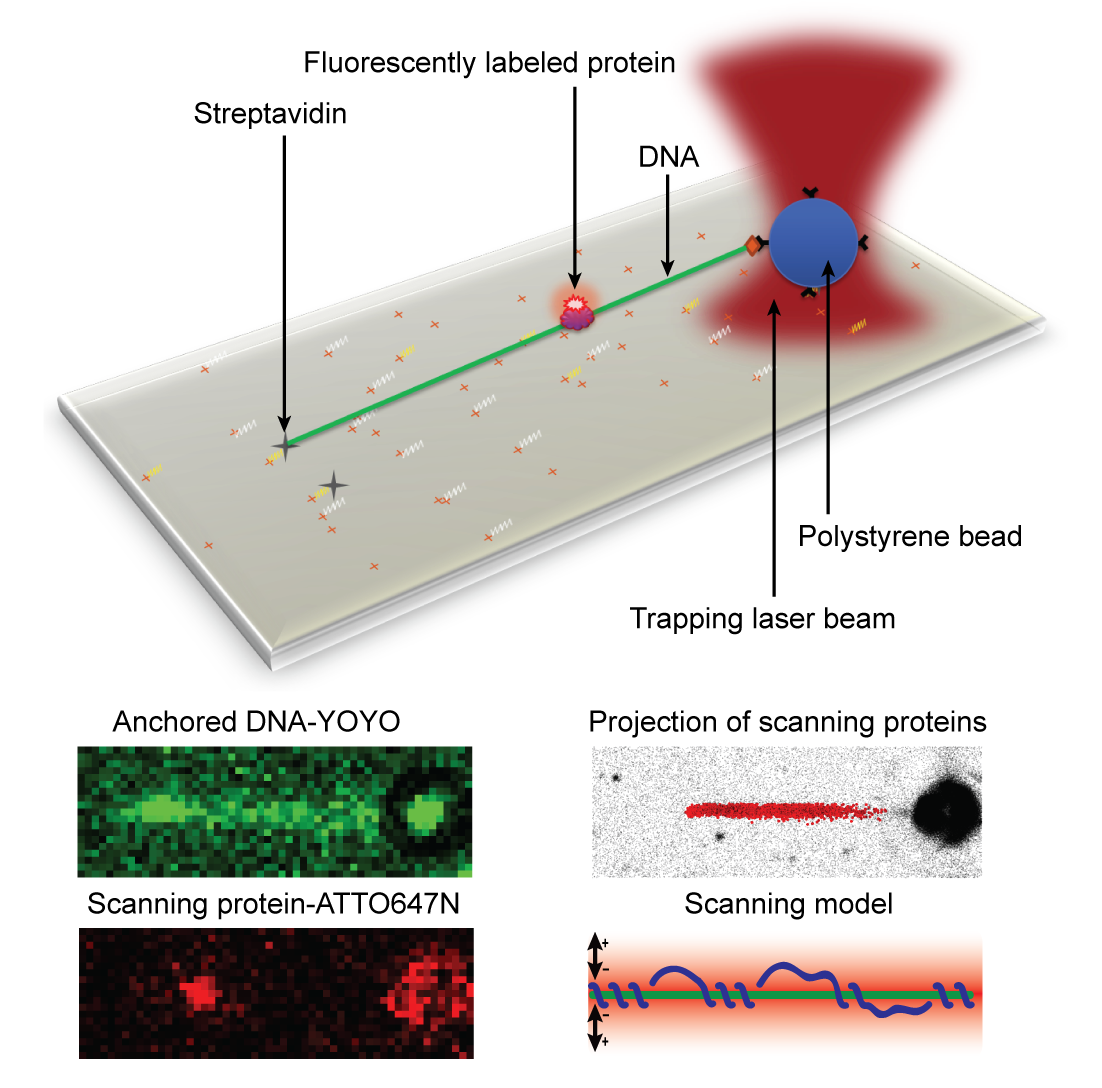
Bottom: The DNA can be visualized using YOYO staining. Single protein molecules can be observed using ATTO647N as a fluorescent probe. The projection of many proteins are collected and analyzed, and a model for how the protein is using a mix between helical ‘spinning’ around the DNA and jumping along the DNA to scan for damages could be construed.
Links:
Breaking the speed limit with multimode fast scanning of DNA by Endonuclease V
Arash Ahmadi, Ida Rosnes, Pernille Blicher, Robin Diekmann, Mark Schüttpelz, Kyrre Glette, Jim Tørresen, Magnar Bjørås, Bjørn Dalhus & Alexander D. Rowe
Nature Communications volume 9, Article number: 5381 (2018)
Home page of the "Structural Biology and DNA repair" research group, led by Bjørn Dalhus
