Researchers at the Department of Immunology publish groundbreaking proteomics technology in Nature Methods.

(click to enlarge image)
The prestigious journal Nature Methods published on October 30th 2018 an article by proteomics researchers at the Department of Immunology at OUH Rikshospitalet. "We describe a simple and affordable method for large-scale protein analysis and test the specificity of 6000 commercially available antibodies to human proteins" says Fridtjof Lund-Johansen who led the study.
Proteins are the building blocks of cells and tissues, and they operate in complex clockworks to exert a host of biological functions. The purpose of large-scale protein analysis, or proteomics, is to dissect these clockworks and understand how they operate. Most drugs act by modifying the function of one or more proteins, so there is good reason to believe that insight from proteomics research will open new avenues for therapy.
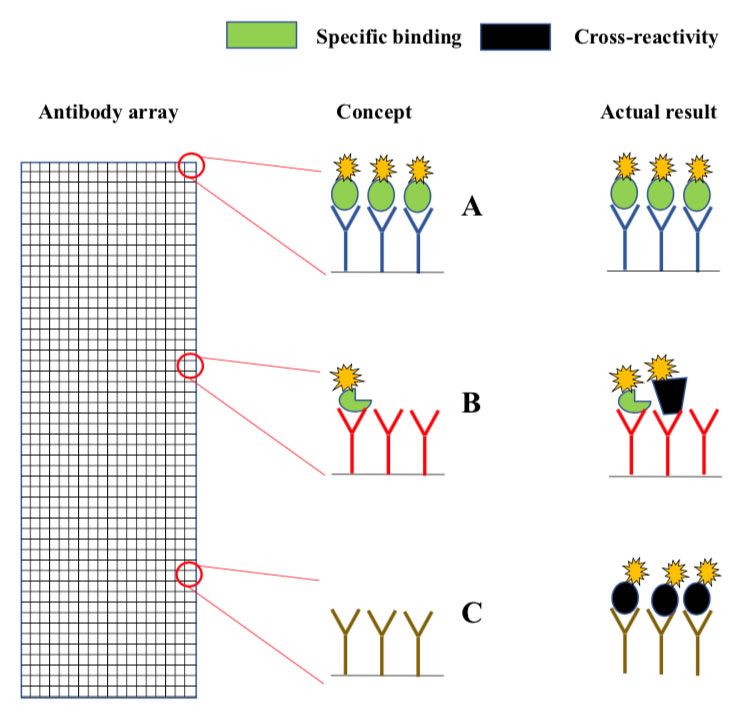
Microarrays are slides analogous to miniature chessboards with thousands of micron-sized squares. Each square contains immobilized antibodies to a different protein (A, B, C). Fluorescently labelled proteins from the sample bind to the antibodies, and a micro-scanner is used to measure fluorescence intensity in each square. Concept: In principle, the fluorescence intensity in each should report on the levels of the antibody target in the sample. Actual result: Since antibodies often cross-react, many of the squares contain a mixture of intended- and non-intended antibody targets (middle), while others may contain only non-intended targets (bottom).(click to enlarge)
An important obstacle in proteomics is that a typical sample occupies a $1M mass spectrometer for an hour or more. The consequence is that very few laboratories can afford to participate in this exciting field of research. To overcome this problem, Lund-Johansen and his team blew new life into a method that was invented nearly twenty years ago but abandoned due to concerns about specificity.
The original method is known as antibody array analysis . The arrays are analogous to miniature chessboards with thousands of micron-sized squares, each coupled with antibodies to a different protein (Fig. 1). The immobilized antibodies capture fluorescently labelled proteins from samples such as serum or cell lysates. The concept is that the fluorescence intensity measured in each square provides information about the levels of the antibody target in the sample. However, since antibodies often cross-react, there is a high chance that many squares contain the “wrong” protein (Fig. 1). Antibody array analysis has therefore long been considered as unreliable.
To solve the problem, the researchers introduced three modifications to the original method. The first was development of arrays that consist of polymer beads with thousands of fluorescent bar codes (Fig. 2). The bead-arrays are analyzed with flow cytometers, which are multi-purpose instruments used by a very large number of researchers. Post-doc Adi Mehta, who produced the arrays for the study, explains that beads have many advantages over slides. “One person can use a robot to produce 3000 bead arrays in a matter of days, and they are very easy to use since all analysis steps are performed in microwell plates”. With large supplies of inexpensive arrays, the researchers had freedom to experiment and think “out of the box”.

Beads with individual color-codes are kept separate until the antibody has been bound. They are then mixed and used to capture fluorescently labelled sample proteins. The beads are analyzed using a flow cytometer that reads the bar codes and measures fluorescence from bound protein. The dotplot shows typical flow cytometry data. The y-and x-axis correspond to one of five colors used to bar-code the beads and fluorescence intensity from bound protein, respectively. Each dot corresponds to an individual bead, and it is common to analyze 30 beads per bar-code. Modern flow cytometers are capable of harvesting samples from microwell plates, and a plate with 96 samples is typically analyzed in 90 minutes.
(click to enlarge)
“A solution to the specificity problem has been right in front of everyone’s eyes since antibody arrays were used the very first time. However, one would need our bead-arrays to think of it”, says Lund-Johansen. He is referring to western blotting, which is the most widely used application for antibodies in research. In this assay, the sample proteins are separated by size before they are labelled with antibodies (Fig. 3, top). As long as the intended target and cross-reactive proteins have different size, they can be detected as discrete bands of antibody staining.
Post-doc Krzysztof Sikorski built on results from early experiments by master student Anette Lie-Jensen to develop an “antibody array western blot”. He separated the labelled sample proteins by size into a series of liquid fractions (Fig. 3, middle). The approach called for as many as 36 bead-arrays to analyze all the fractions, but as a result, the researchers could resolve different targets binding to each of 6000 antibodies as peaks of reactivity across the fractions (Fig. 3, upper line chart). However, there was still need for a method to identify the peaks that correspond to the intended antibody targets.
The missing piece of the puzzle came into place through a collaboration with proteomics researchers Gustavo de Souza, Maria Ekmann-Stensland and Tuula Nyman. They helped Sikorski use the highly advanced mass spectrometers in the OUS proteomics core facility. Mass spectrometry analysis provided a detailed map over the distribution of thousands of proteins in the fractions (Fig. 3 bottom). Thus, the reactivity peaks for each antibody was compared to the peak identified by mass spectrometry (Fig. 3, line charts). This paved the way for the automated analysis of antibody specificity described in the Nature Methods article. Once the antibodies have been validated, the “antibody array western blot” can be used as a stand-alone method independently of mass spectrometry.
The researchers have nicknamed their technology “personal proteomics” to allude to the fact that nearly everyone can use it. “The method is simpler to perform than standard western blotting” says Lund-Johansen. As head of the flow cytometry core facility of OUH Rikshospitalet, he can safely say that there is considerable instrument capacity. The group’s next task is to identify useful antibodies against everyone’s favorite proteins. The researchers collaborate with a large number of antibody manufacturers, but they also hope for a community effort, so anyone who wish to have their antibodies tested may contact them. “A few micrograms of antibody is sufficient”, says Lund-Johansen.
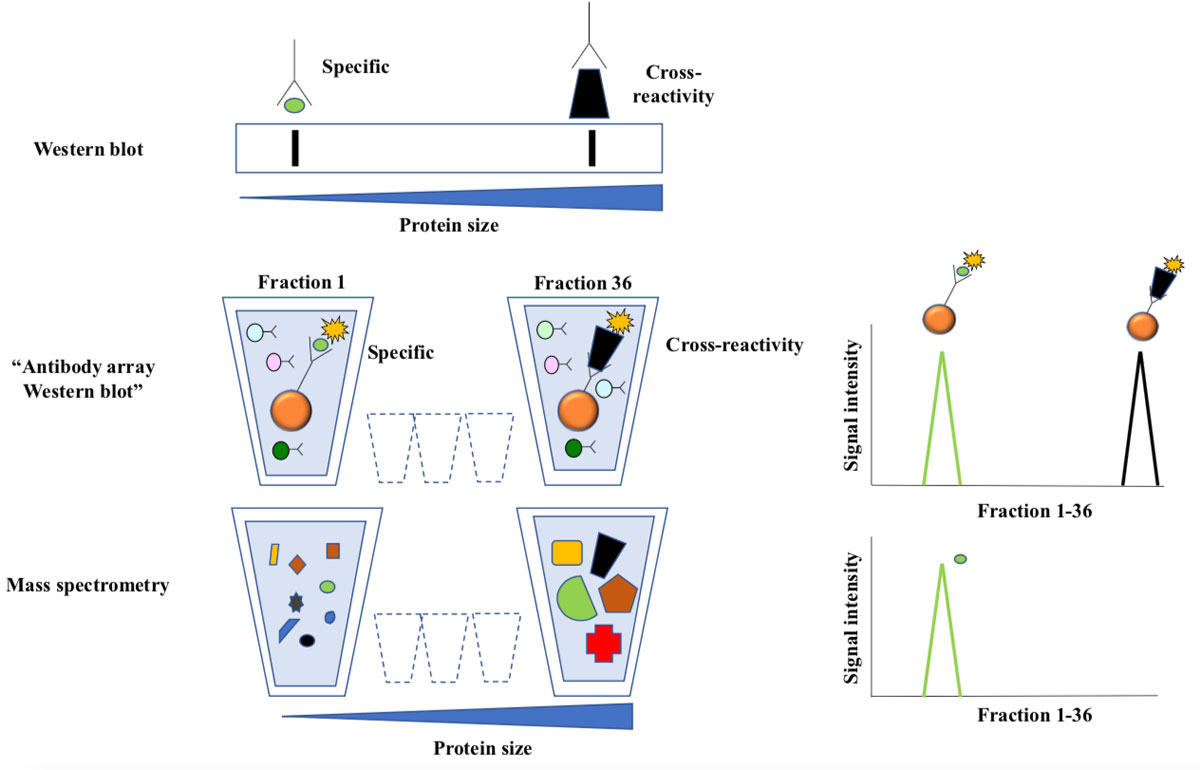
Figure 3 Antibody array western blotting.
Top: In traditional western blotting antibodies are used to detect size-separated proteins immobilized to a membrane. Antibody staining is visualized as bands, and the position of the band corresponding to the intended target is predictable from size of the intended antibody target. Extra bands represent cross-reactivity.
Middle: Labelled proteins from cell lysates were separated according to size by preparative gel-electrophoresis. The approach yields 36 liquid fractions, each containing proteins with a narrow size distribution. Antibody bead-arrays were added to each fraction and analyzed by flow cytometry. The line chart illustrates how two targets of the same antibody are resolved as peaks of reactivity across the fractions.
The bottom line chart shows distribution of the intended target as measured by mass spectrometry. This technique allows parallel analysis of thousands of proteins in each sample, and the results are very reliable since the detection is based on protein-sequencing.
(click to enlarge)
Testing antibody specificity is probably a wise thing to do. Over the past years, feature articles and editorials in Nature have described what is referred to as a reproducibility crisis in bio-medical research, and poorly validated antibodies top the list of villains 2-5. In fact, some of the articles forecast the paper published this week, and describe the technology as a possible solution to current problems 2, 3.
The researchers wish to acknowledge the generous support from the KG Jebsen Foundation and their colleagues in the KG Jebsen Centre for Cancer Immunotherapy. The work was also supported by innovation grants from Helse-Sør-Øst, The Norwegian Research Council and Novo-Nordisk Foundation.
Media attention:
The major national news outlet for health and medicine "Dagens Medisin" has covered the story.
References:
- Sikorski, K., Mehta, A., Inngjerdingen, M., Thakor, F., Kling, S., Kalina, T., Nyman, T.A., Stensland, M., De Souza, G.A., Holden, L., Stuchly, J., Templin, M. & Lund-Johansen, F.
A high throughput pipeline for validation of antibodies.
Nature Methods (2018). Published: 30 October 2018
Open article - Baker, M. Antibody anarchy: A call to order. Nature 527, 545-551 (2015).
- Baker, M. Blame it on the antibodies. Nature 521, 274-276 (2015).
- Bradbury, A. & Pluckthun, A. Reproducibility: Standardize antibodies used in research. Nature 518, 27-29 (2015).
- Marx, V. Finding the right antibody for the job. Nat. Methods 10, 14 (2013).
Links:
New article from Dagens Medisin (in Norwegian):
OUS-forskere står bak ny testmetode for antistoffer
Home page of Fridtjof Lund-Johansen's Protein array group
Home page of Marit Inngjerdingen's Innate lymphocyte group
Previous news article from ous-research.no:
OUH researchers publish groundbreaking computer tools for cell biology research in Nature Methods (29.08.2016)
