Alternate transcripts from a single gene locus greatly enhance the combinatorial flexibility of the human transcriptome. Different patterns of exon usage have been observed when comparing normal tissue to cancers, suggesting that variant transcripts may play a role in the tumor phenotype.
Subtype and cell type specific expression of lncRNAs provide insight into breast cancer
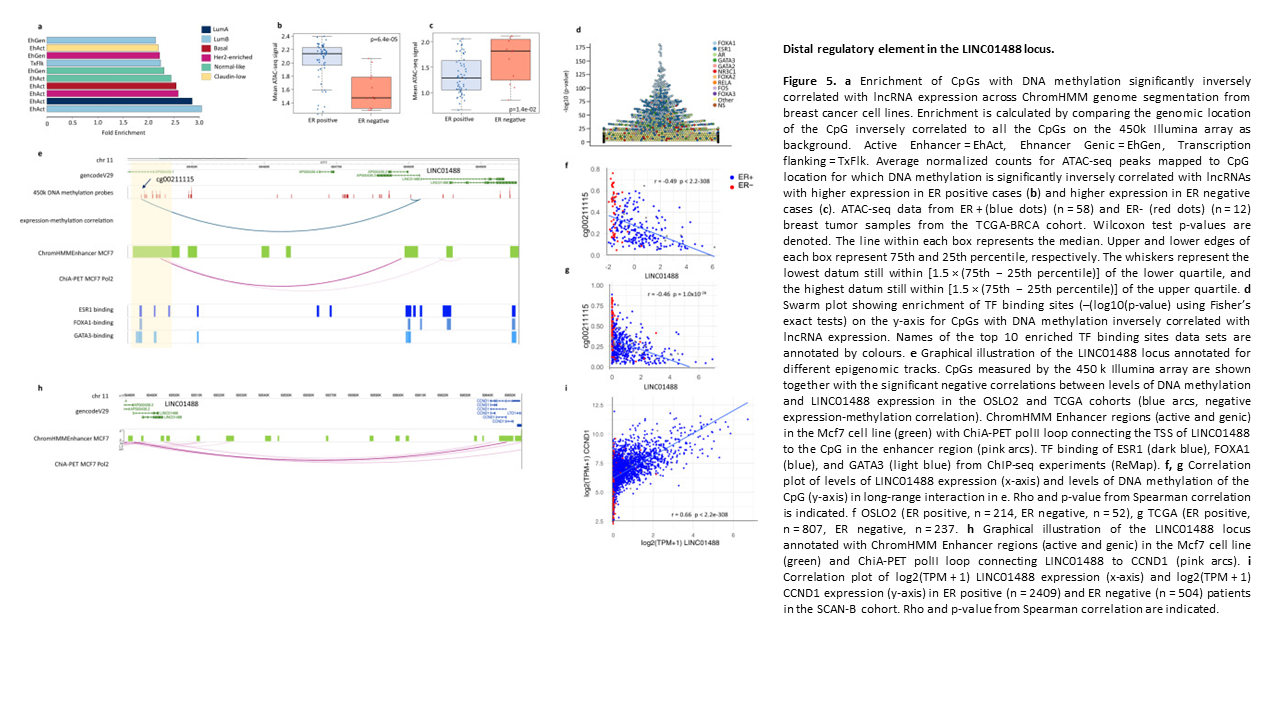
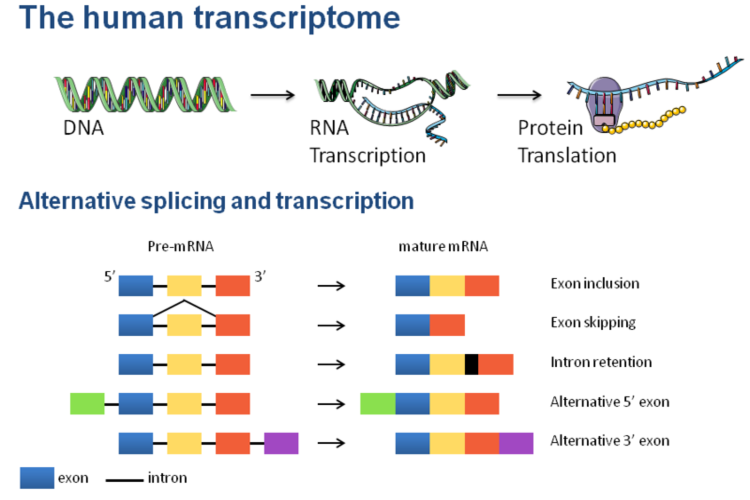
Figure 1.The human transcriptome.
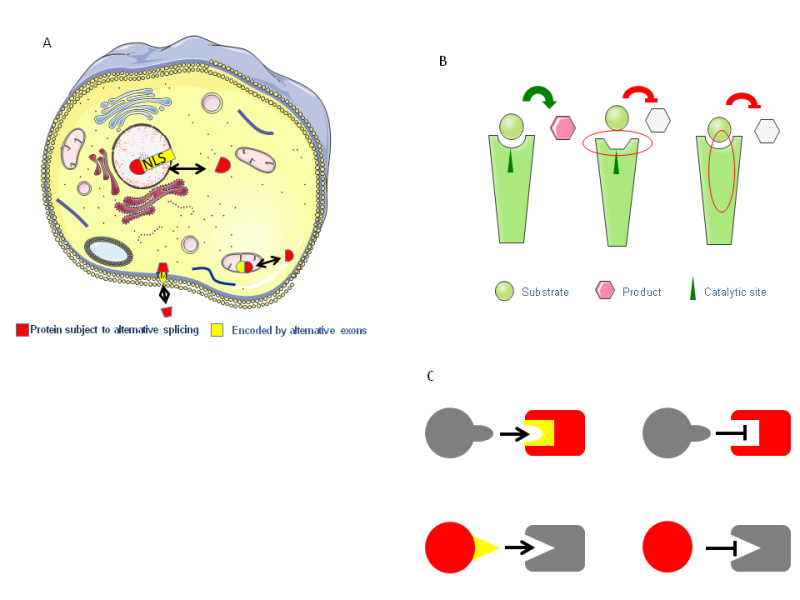
Figure 2.Protein diversity as a result of alternative splicing and transcription.The effect of alternative splicing can range from no effect at all, to complete degradation. A)But many times an exon can represent a distinct protein domain, and can influence protein function by changing the cellular localization. B)If the protein is an enzyme the enzymatic activity could be altered by changing the binding of substrates or catalytic domain. C) It can change the binding properties of proteins by affecting the binding pocket or the binding domain.
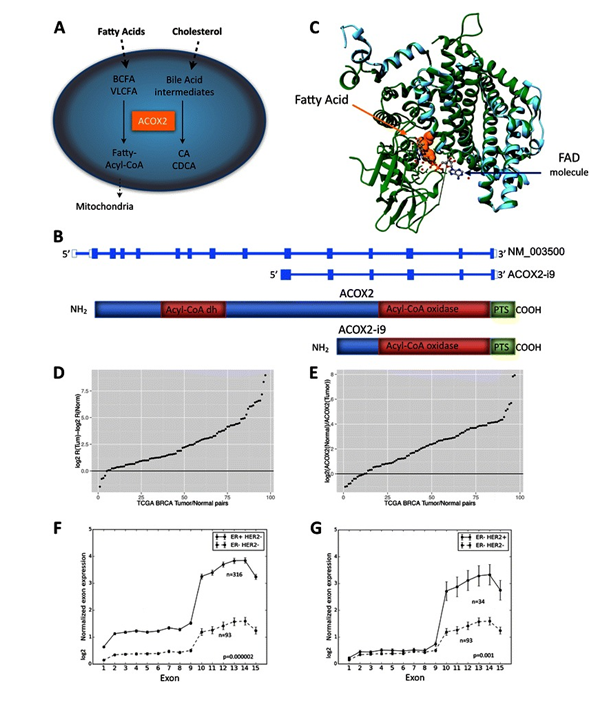
Figure 3.ACOX2 expression in TCGA BRCA cohort. ACOX2 is involved in the oxidation of very long chain fatty acids, VLCFA, and branched chain fatty acids, BCFA, and in the synthesis of bile-acid precursor molecules as schematically illustrated in Fig. 1a. The ACOX2 intronic variant, ACOX2-i9, is initiated just upstream of exon 10 of the full-length transcript (b). The translated protein retains the Acyl-CoA oxidase domain, and the Preoxisomal Targeting Signal, but lacks the fatty acid binding domain of the full-length protein. 1c shows a model of ACOX2-i9 aligned with 2DDH (Rat ACOX2). The template 2DDH is colored green and the model (i9) is colored cyan. The fatty acid is depicted as orange colored spheres and the FAD molecule (and water depicted as small red spheres) is depicted as ball-stick and colored by element. The difference in Log2 R(ACOX2-i9/ACOX2) Tumor – log2 R Normal in 87 tumor/normal pairs from the TCGA BRCA dataset are shown in d. Values > 0 indicate that the Ratio of ACOX2-i9/ACOX2 is higher in the tumor. e shows log2 expression of ACOX2 in Normal/Tumor. ACOX2 is expressed at higher levels in the Normal sample when the log2 ratio >0. Normalized log2 RPKM expression of each exon of ACOX2 in Her2 negative background separated by ER status are shown in f, and in ER negative patients separated by Her2 status (g). FromExpression of an estrogen-regulated variant transcript of the peroxisomal branched chain fatty acid oxidase ACOX2 in breast carcinomas. BMC Cancer. 2015 Jul 17;15:524. doi: 10.1186/s12885-015-1510-8. PMID:26183823
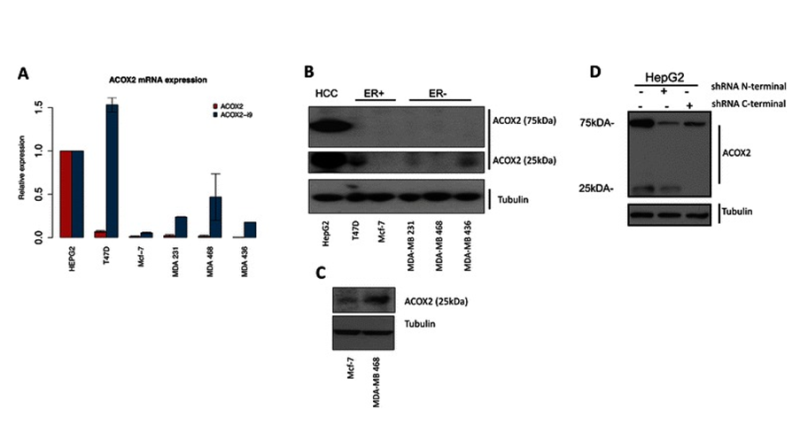
Figure 4.Expression of ACOX2 in breast cancer cell lines. ACOX2 and ACOX2-i9 mRNA levels were assessed by qRT-PCR in HepG2 cells and breast cancer cell lines (a), quantification is shown relative to HepG2 expression. Protein extracts from HepG2 and breast cancer cell lines were probed with a C-terminal antibody against ACOX2 (b). Highly sensitive chemiluminescent substrate (c) was included for illustration purposes to show even low levels of protein expression. HepG2 cells were transfected with shRNA targeting the N-terminal and C-terminal regions of ACOX2 (d). From Expression of an estrogen-regulated variant transcript of the peroxisomal branched chain fatty acid oxidase ACOX2 in breast carcinomas. BMC Cancer. 2015 Jul 17;15:524. doi: 10.1186/s12885-015-1510-8. PMID:26183823
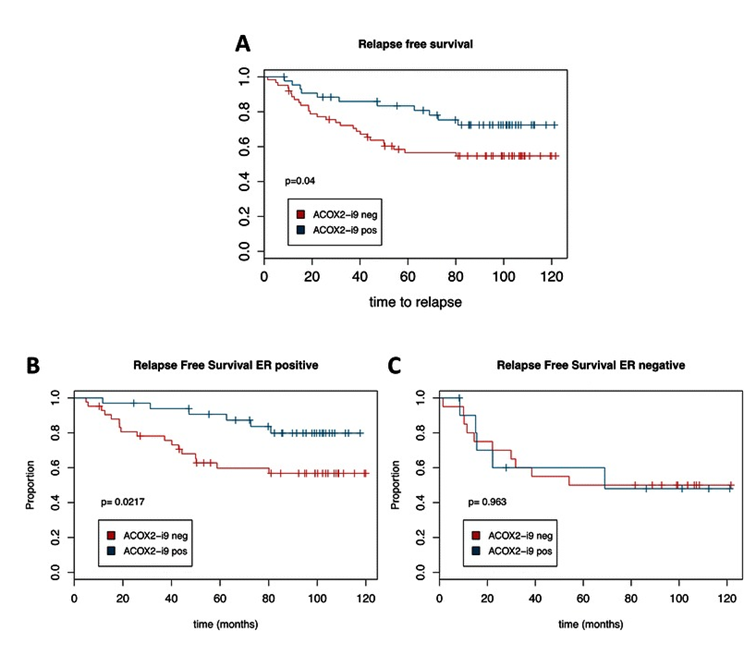
Figure 5.ACOX2-i9 expression is associated with good prognosis in a cohort of breast cancer patients. Kaplan-Meier survival curves of patients from the MicMa cohort testing positive (n = 44) or negative (n = 62) for ACOX2-i9 by PCR assay (a). b and c show survival curves for ER positive (ACOX2-i9pos n = 33, ACOX2-i9neg n = 42) and ER negative patients (ACOX2-i9pos n = 11, ACOX2-i9neg n = 20) respectively. From Expression of an estrogen-regulated variant transcript of the peroxisomal branched chain fatty acid oxidase ACOX2 in breast carcinomas. BMC Cancer. 2015 Jul 17;15:524. doi: 10.1186/s12885-015-1510-8. PMID:26183823
Project members: Sunniva Bjørklund, Grethe I. Grenaker Alnæs
PhD thesis from project:
Sunniva Bjørklund 08.06.2016; Title of the thesis: Next- generation sequencing of non-canonical biological systems in breast cancer.
External colaborators:
- Gyan Bhanot, Rutgers University, NJ, USA http://evolution.rutgers.edu/people/ches-faculty-mainmenu-150/test
- Shridar Ganesan, Cancer Institute, NJ, USA https://www.cinj.org/shridar-ganesan-md-phd
- Ravi Shachaninandam, Mt Sinay School of medicine http://www.mountsinai.org/profiles/ravi-sachidanandam
Recent publications:
1. Expression of an estrogen-regulated variant transcript of the peroxisomal branched chain fatty acid oxidase ACOX2 in breast carcinomas.
BMC Cancer. 2015 Jul 17;15:524. doi: 10.1186/s12885-015-1510-8. PMID:26183823
2. Subtype and cell type specific expression of lncRNAs provide insight into breast cancer
Commun Biol. 2022; 5: 834. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9388662/