Goal of our yeast work
Cells are continuously exposed to changes in their environment. This is not only a challenge for single-celled organisms like budding yeast, but it also holds true for for example tumor cells, which can be exposed to various forms of stress including low oxygen levels, chemotherapy, radiation, and low nutrient levels. For optimal growth and survival, cells have developed mechanisms that sense environmental alterations and generate responses that maintain cellular homeostasis. A major response to environmental stimuli is transcriptional reprogramming. We use the budding yeast Saccharomyces cerevisiae as a model organism to understand how cells cope with environmental changes, in particular in terms of regulation of transcription. We mainly focus on regulation of transcription by post-translational modifications, such as phosphorylation, ubiquitination and sumoylation.
1. Sumoylation and cell stress
We want to understand how cells modulate transcription in response to environmental changes, in particular nutrient starvation. We are particularly interested in the role of the ubiquitin family protein Sumo. Sumoylation is clearly important in rewiring of transcription in response to cell stress. However, which proteins are sumoylated and how Sumo regulates their function remains mysterious. By performing ChIP-seq experiments and mass spectrometry we have identified a number of proteins that become sumoylated during cell stress. We are currently studying the molecular mechanism by which Sumo modulates the activity of these proteins to regulate transcription.
2. Regulation of transcription by signal transduction kinases and phosphatases
Another major focus of the group is to determine how kinases and phosphatases regulate transcription. In response to specific environmental cues, signaling pathways rewire transcriptional programs to make sure the cell optimally responds to changes in conditions. These signaling pathways often consist of kinases and phosphatases. However, how these enzymes control transcription is not well understood. Using a combination of ChIP-seq, mass spectrometry and fluorescence microscopy we are unraveling the molecular mechanisms by which signaling kinases and phosphatases control transcription.
Previous findings
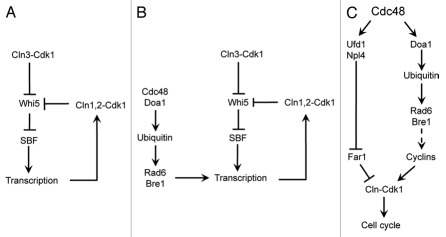
Rad6 in transcription and cell cycle regulation
Using a high-throughput chemical-genetic screen, we found that the E2 ubiquitin conjugase Rad6 is temporally recruited to promoter regions of cyclin genes, where it ubiquitinates histone H2B (Fig. 1). This promotes transcription of cyclins, thereby activating the cyclin dependent kinase Cdk1 to promote efficient cell cycle entry.
Cdk1 in regulation of transcription
We discovered that Cdk1 has a kinase-dependent function in regulation of the basal transcription machinery. Using ChIP-seq, we found that Cdk1 localizes to highly transcribed genes, primarily involved in housekeeping. At these genes, Cdk1 cooperates with another CDK, Kin28 (the catalytic subunit of TFIIH), to phosphorylate RNA polymerase II (Fig. 2). RNA polymerase II has a long C-terminal domain that consists of 28 repeats of the sequence Y1S2P3T4S5P6S7. Cdk1 and Kin28 promote phosphorylation of the serine residue at position 5 (Ser5). This in turn is important for efficient recruitment of the capping machinery, which places a cap on the mRNA as soon as it protrudes from the polymerase.
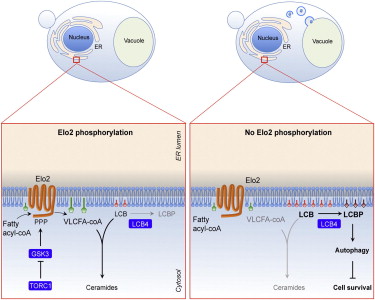
Cell reponses to nutrient stress: Regulation of fatty acid and ceramide metabolism by TORC1 and GSK3
Very long chain fatty acids (VLCFAs) are essential fatty acids with multiple functions, including ceramide synthesis. Although the components of the VLCFA biosynthetic machinery have been elucidated, how their activity is regulated to meet the cell’s metabolic demand remains unknown. We recently found that the fatty acid elongase Elo2 is regulated by phosphorylation. Elo2 phosphorylation is induced by nutrient stress and inhibition of TORC1 and requires GSK3. We found that expression of nonphosphorylatable Elo2 profoundly altered the ceramide spectrum, which reflects aberrant VLCFA synthesis. Furthermore, depletion of VLCFAs resulted in constitutive activation of autophagy. This constitutive activation of autophagy diminished cell survival, indicating that VLCFAs serve to dampen the amplitude of autophagy. Together, our data revealed a function for TORC1 and GSK3 in the regulation of VLCFA synthesis with important implications for autophagy and cell homeostasis.